Structural insights into melatonin receptors
- PMID: 31693784
- PMCID: PMC7174090
- DOI: 10.1111/febs.15128
Structural insights into melatonin receptors
Abstract
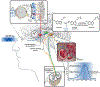
The long-anticipated high-resolution structures of the human melatonin G protein-coupled receptors MT1 and MT2 , involved in establishing and maintaining circadian rhythm, were obtained in complex with two melatonin analogs and two approved anti-insomnia and antidepression drugs using X-ray free-electron laser serial femtosecond crystallography. The structures shed light on the overall conformation and unusual structural features of melatonin receptors, as well as their ligand binding sites and the melatonergic pharmacophore, thereby providing insights into receptor subtype selectivity. The structures revealed an occluded orthosteric ligand binding site with a membrane-buried channel for ligand entry in both receptors, and an additional putative ligand entry path in MT2 from the extracellular side. This unexpected ligand entry mode contributes to facilitating the high specificity with which melatonin receptors bind their cognate ligand and exclude structurally similar molecules such as serotonin, the biosynthetic precursor of melatonin. Finally, the MT2 structure allowed accurate mapping of type 2 diabetes-related single-nucleotide polymorphisms, where a clustering of residues in helices I and II on the protein-membrane interface was observed which could potentially influence receptor oligomerization. The role of receptor oligomerization is further discussed in light of the differential interaction of MT1 and MT2 with GPR50, a regulatory melatonin coreceptor. The melatonin receptor structures will facilitate design of selective tool compounds to further dissect the specific physiological function of each receptor subtype as well as provide a structural basis for next-generation sleeping aids and other drugs targeting these receptors with higher specificity and fewer side effects.
Keywords: G protein-coupled receptors; X-ray free-electron laser; chronobiology; diabetes; melatonin; serial femtosecond crystallography; serotonin; sleeping aids.
© 2019 Federation of European Biochemical Societies.
Conflict of interest statement
Conflict of interest
The authors declare no competing interests.
Figures

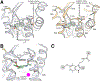


Similar articles
-
XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity.Nature. 2019 May;569(7755):289-292. doi: 10.1038/s41586-019-1144-0. Epub 2019 Apr 24. Nature. 2019. PMID: 31019305 Free PMC article.
-
A molecular and chemical perspective in defining melatonin receptor subtype selectivity.Int J Mol Sci. 2013 Sep 6;14(9):18385-406. doi: 10.3390/ijms140918385. Int J Mol Sci. 2013. PMID: 24018885 Free PMC article. Review.
-
Structural basis of the ligand binding and signaling mechanism of melatonin receptors.Nat Commun. 2022 Jan 24;13(1):454. doi: 10.1038/s41467-022-28111-3. Nat Commun. 2022. PMID: 35075127 Free PMC article.
-
Molecular basis of ligand selectivity for melatonin receptors.RSC Adv. 2023 Feb 1;13(7):4422-4430. doi: 10.1039/d2ra06693a. eCollection 2023 Jan 31. RSC Adv. 2023. PMID: 36760312 Free PMC article.
-
Melatonin Receptor Signaling: Impact of Receptor Oligomerization on Receptor Function.Int Rev Cell Mol Biol. 2018;338:59-77. doi: 10.1016/bs.ircmb.2018.02.002. Epub 2018 Apr 5. Int Rev Cell Mol Biol. 2018. PMID: 29699692 Review.
Cited by
-
Empowering Melatonin Therapeutics with Drosophila Models.Diseases. 2021 Sep 26;9(4):67. doi: 10.3390/diseases9040067. Diseases. 2021. PMID: 34698120 Free PMC article. Review.
-
Melatonin's Impact on Wound Healing.Antioxidants (Basel). 2024 Oct 2;13(10):1197. doi: 10.3390/antiox13101197. Antioxidants (Basel). 2024. PMID: 39456451 Free PMC article. Review.
-
Melatonin alleviates septic ARDS by inhibiting NCOA4-mediated ferritinophagy in alveolar macrophages.Cell Death Discov. 2024 May 24;10(1):253. doi: 10.1038/s41420-024-01991-8. Cell Death Discov. 2024. PMID: 38789436 Free PMC article.
-
Melatonin Regulates Lipid Metabolism in Porcine Cumulus-Oocyte Complexes via the Melatonin Receptor 2.Antioxidants (Basel). 2022 Mar 31;11(4):687. doi: 10.3390/antiox11040687. Antioxidants (Basel). 2022. PMID: 35453372 Free PMC article.
-
Utility of melatonin in mitigating ionizing radiation-induced testis injury through synergistic interdependence of its biological properties.Biol Res. 2022 Nov 4;55(1):33. doi: 10.1186/s40659-022-00401-6. Biol Res. 2022. PMID: 36333811 Free PMC article.
References
-
- Aschoff J (1965) Circadian Rhythms in Man. Science 148, 1427–1432. - PubMed
-
- Cipolla-Neto J & Amaral FGD (2018) Melatonin as a Hormone: New Physiological and Clinical Insights. Endocr Rev 39, 990–1028. - PubMed
-
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M & Hashimoto S (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks, Nat Genet 37, 187–192. - PubMed
-
- Aschoff J (1981) Freerunning and Entrained Circadian Rhythms in Biological Rhythms (Aschoff J, ed) pp. 81–93, Springer US, Boston, MA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

