Hox11 expressing regional skeletal stem cells are progenitors for osteoblasts, chondrocytes and adipocytes throughout life
- PMID: 31320650
- PMCID: PMC6639390
- DOI: 10.1038/s41467-019-11100-4
Hox11 expressing regional skeletal stem cells are progenitors for osteoblasts, chondrocytes and adipocytes throughout life
Abstract
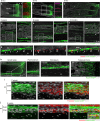
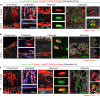
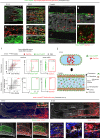
Multipotent mesenchymal stromal cells (MSCs) are required for skeletal formation, maintenance, and repair throughout life; however, current models posit that postnatally arising long-lived adult MSCs replace transient embryonic progenitor populations. We previously reported exclusive expression and function of the embryonic patterning transcription factor, Hoxa11, in adult skeletal progenitor-enriched MSCs. Here, using a newly generated Hoxa11-CreERT2 lineage-tracing system, we show Hoxa11-lineage marked cells give rise to all skeletal lineages throughout the life of the animal and persist as MSCs. Hoxa11 lineage-positive cells give rise to previously described progenitor-enriched MSC populations marked by LepR-Cre and Osx-CreER, placing them upstream of these populations. Our studies establish that Hox-expressing cells are skeletal stem cells that arise from the earliest stages of skeletal development and self-renew throughout the life of the animal.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Novel Lineage-Tracing System to Identify Site-Specific Ectopic Bone Precursor Cells.Stem Cell Reports. 2021 Mar 9;16(3):626-640. doi: 10.1016/j.stemcr.2021.01.011. Epub 2021 Feb 18. Stem Cell Reports. 2021. PMID: 33606989 Free PMC article.
-
Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow.Cell Stem Cell. 2014 Aug 7;15(2):154-68. doi: 10.1016/j.stem.2014.06.008. Epub 2014 Jun 19. Cell Stem Cell. 2014. PMID: 24953181 Free PMC article.
-
Hes1 marks peri-condensation mesenchymal cells that generate both chondrocytes and perichondrial cells in early bone development.J Biol Chem. 2023 Jun;299(6):104805. doi: 10.1016/j.jbc.2023.104805. Epub 2023 May 11. J Biol Chem. 2023. PMID: 37172728 Free PMC article.
-
Bone marrow mesenchymal stem cells: fat on and blast off by FGF21.Int J Biochem Cell Biol. 2013 Mar;45(3):546-9. doi: 10.1016/j.biocel.2012.12.014. Epub 2012 Dec 25. Int J Biochem Cell Biol. 2013. PMID: 23270727 Free PMC article. Review.
-
Application of Mesenchymal Stem Cells in Inflammatory and Fibrotic Diseases.Int J Mol Sci. 2020 Nov 7;21(21):8366. doi: 10.3390/ijms21218366. Int J Mol Sci. 2020. PMID: 33171878 Free PMC article. Review.
Cited by
-
MYC promotes fibroblast osteogenesis by regulating ALP and BMP2 to participate in ectopic ossification of ankylosing spondylitis.Arthritis Res Ther. 2023 Feb 21;25(1):28. doi: 10.1186/s13075-023-03011-z. Arthritis Res Ther. 2023. PMID: 36803548 Free PMC article.
-
Endothelial and Leptin Receptor+ cells promote the maintenance of stem cells and hematopoiesis in early postnatal murine bone marrow.Dev Cell. 2023 Mar 13;58(5):348-360.e6. doi: 10.1016/j.devcel.2023.02.003. Epub 2023 Mar 2. Dev Cell. 2023. PMID: 36868235 Free PMC article.
-
Periosteal Skeletal Stem and Progenitor Cells in Bone Regeneration.Curr Osteoporos Rep. 2022 Oct;20(5):334-343. doi: 10.1007/s11914-022-00737-8. Epub 2022 Jul 13. Curr Osteoporos Rep. 2022. PMID: 35829950 Review.
-
Transcriptomic analysis of bone and fibrous tissue morphogenesis during digit tip regeneration in the adult mouse.FASEB J. 2020 Jul;34(7):9740-9754. doi: 10.1096/fj.202000330R. Epub 2020 Jun 7. FASEB J. 2020. PMID: 32506623 Free PMC article.
-
Novel Lineage-Tracing System to Identify Site-Specific Ectopic Bone Precursor Cells.Stem Cell Reports. 2021 Mar 9;16(3):626-640. doi: 10.1016/j.stemcr.2021.01.011. Epub 2021 Feb 18. Stem Cell Reports. 2021. PMID: 33606989 Free PMC article.
References
-
- Rux Danielle R., Song Jane Y., Swinehart Ilea T., Pineault Kyriel M., Schlientz Aleesa J., Trulik Kelsey G., Goldstein Steve A., Kozloff Ken M., Lucas Daniel, Wellik Deneen M. Regionally Restricted Hox Function in Adult Bone Marrow Multipotent Mesenchymal Stem/Stromal Cells. Developmental Cell. 2016;39(6):653–666. doi: 10.1016/j.devcel.2016.11.008. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

