Contribution of DNA methylation to the expression of FCGRT in human liver and myocardium
- PMID: 31209240
- PMCID: PMC6572836
- DOI: 10.1038/s41598-019-45203-1
Contribution of DNA methylation to the expression of FCGRT in human liver and myocardium
Abstract
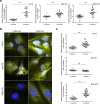
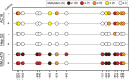
FcRn mediates recycling and transcytosis of IgG and albumin in various cell types. The MHC-class-I-like protein of the FcRn heterodimer is encoded by FCGRT. Few determinants of variable FCGRT expression in humans have been identified so far. In this study, we investigated the presence of DNA methylation in regulatory regions of FCGRT in samples of human liver and myocardium tissue, and we examined the impact of FCGRT methylation on FcRn expression in model cell lines. Quantitative DNA methylation analysis of the FCGRT locus revealed differentially methylated regions in DNA from liver and myocardium. Methylation status in individual CpG sites correlated with FCGRT mRNA expression. Data from model cell lines suggest that differential methylation in the -1058 to -587 bp regulatory region of FCGRT contributes to FcRn expression. Chromatin immunoprecipitation assays indicate that CpG site methylation impacts the binding of the methylation sensitive transcription factors Zbtb7a and Sp1. This study provides a foundation to further define the contribution of epigenetic factors during the control of FcRn expression and IgG traffic in human tissues.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Regulation of the Human Fc-Neonatal Receptor alpha-Chain Gene FCGRT by MicroRNA-3181.Pharm Res. 2018 Jan 4;35(1):15. doi: 10.1007/s11095-017-2294-0. Pharm Res. 2018. PMID: 29302759 Free PMC article.
-
Analysis of Response Elements Involved in the Regulation of the Human Neonatal Fc Receptor Gene (FCGRT).PLoS One. 2015 Aug 7;10(8):e0135141. doi: 10.1371/journal.pone.0135141. eCollection 2015. PLoS One. 2015. PMID: 26252948 Free PMC article.
-
Hepatic FcRn regulates albumin homeostasis and susceptibility to liver injury.Proc Natl Acad Sci U S A. 2017 Apr 4;114(14):E2862-E2871. doi: 10.1073/pnas.1618291114. Epub 2017 Mar 22. Proc Natl Acad Sci U S A. 2017. PMID: 28330995 Free PMC article.
-
The neonatal Fc receptor in cancer FcRn in cancer.Cancer Med. 2020 Jul;9(13):4736-4742. doi: 10.1002/cam4.3067. Epub 2020 May 5. Cancer Med. 2020. PMID: 32368865 Free PMC article. Review.
-
The multiple facets of FcRn in immunity.Immunol Rev. 2015 Nov;268(1):253-68. doi: 10.1111/imr.12331. Immunol Rev. 2015. PMID: 26497526 Review.
Cited by
-
Receptors and Host Factors for Enterovirus Infection: Implications for Cancer Therapy.Cancers (Basel). 2024 Sep 12;16(18):3139. doi: 10.3390/cancers16183139. Cancers (Basel). 2024. PMID: 39335111 Free PMC article. Review.
-
Albumin influences leucocyte FcRn expression in the early days of kidney transplantation.Clin Exp Immunol. 2024 May 16;216(3):307-317. doi: 10.1093/cei/uxae011. Clin Exp Immunol. 2024. PMID: 38353127 Free PMC article.
-
The therapeutic age of the neonatal Fc receptor.Nat Rev Immunol. 2023 Jul;23(7):415-432. doi: 10.1038/s41577-022-00821-1. Epub 2023 Feb 1. Nat Rev Immunol. 2023. PMID: 36726033 Free PMC article. Review.
-
The neonatal Fc receptor expression during macrophage differentiation is related to autophagy.Front Immunol. 2022 Nov 1;13:1054425. doi: 10.3389/fimmu.2022.1054425. eCollection 2022. Front Immunol. 2022. PMID: 36389739 Free PMC article.
-
Analysis of the intracellular traffic of IgG in the context of Down syndrome (trisomy 21).Sci Rep. 2021 May 26;11(1):10981. doi: 10.1038/s41598-021-90469-z. Sci Rep. 2021. PMID: 34040082 Free PMC article.
References
-
- Praetor A, Hunziker W. beta(2)-Microglobulin is important for cell surface expression and pH-dependent IgG binding of human FcRn. J Cell Sci. 2002;115:2389–2397. - PubMed
-
- Antohe, F., Radulescu, L., Gafencu, A., Ghetie, V. & Simionescu, M. Expression of functionally active FcRn and the differentiated bidirectional transport of IgG in human placental endothelial cells. Hum Immunol62, 93–105, doi:S0198885900002445 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

