Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2
- PMID: 30102747
- PMCID: PMC6107290
- DOI: 10.1371/journal.ppat.1007236
Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2
Abstract
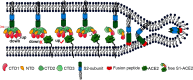
The trimeric SARS coronavirus (SARS-CoV) surface spike (S) glycoprotein consisting of three S1-S2 heterodimers binds the cellular receptor angiotensin-converting enzyme 2 (ACE2) and mediates fusion of the viral and cellular membranes through a pre- to postfusion conformation transition. Here, we report the structure of the SARS-CoV S glycoprotein in complex with its host cell receptor ACE2 revealed by cryo-electron microscopy (cryo-EM). The complex structure shows that only one receptor-binding domain of the trimeric S glycoprotein binds ACE2 and adopts a protruding "up" conformation. In addition, we studied the structures of the SARS-CoV S glycoprotein and its complexes with ACE2 in different in vitro conditions, which may mimic different conformational states of the S glycoprotein during virus entry. Disassociation of the S1-ACE2 complex from some of the prefusion spikes was observed and characterized. We also characterized the rosette-like structures of the clustered SARS-CoV S2 trimers in the postfusion state observed on electron micrographs. Structural comparisons suggested that the SARS-CoV S glycoprotein retains a prefusion architecture after trypsin cleavage into the S1 and S2 subunits and acidic pH treatment. However, binding to the receptor opens up the receptor-binding domain of S1, which could promote the release of the S1-ACE2 complex and S1 monomers from the prefusion spike and trigger the pre- to postfusion conformational transition.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.Sci Rep. 2018 Oct 24;8(1):15701. doi: 10.1038/s41598-018-34171-7. Sci Rep. 2018. PMID: 30356097 Free PMC article.
-
Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein.Cell. 2020 Apr 16;181(2):281-292.e6. doi: 10.1016/j.cell.2020.02.058. Epub 2020 Mar 9. Cell. 2020. PMID: 32155444 Free PMC article.
-
Role of the GTNGTKR motif in the N-terminal receptor-binding domain of the SARS-CoV-2 spike protein.Virus Res. 2020 Sep;286:198058. doi: 10.1016/j.virusres.2020.198058. Epub 2020 Jun 9. Virus Res. 2020. PMID: 32531235 Free PMC article.
-
Mass Spectrometry and Structural Biology Techniques in the Studies on the Coronavirus-Receptor Interaction.Molecules. 2020 Sep 10;25(18):4133. doi: 10.3390/molecules25184133. Molecules. 2020. PMID: 32927621 Free PMC article. Review.
-
Receptor recognition and cross-species infections of SARS coronavirus.Antiviral Res. 2013 Oct;100(1):246-54. doi: 10.1016/j.antiviral.2013.08.014. Epub 2013 Aug 29. Antiviral Res. 2013. PMID: 23994189 Free PMC article. Review.
Cited by
-
Comparison of the Neutralization Power of Sotrovimab Against SARS-CoV-2 Variants: Development of a Rapid Computational Method.JMIR Bioinform Biotechnol. 2024 Oct 10;5:e58018. doi: 10.2196/58018. JMIR Bioinform Biotechnol. 2024. PMID: 39388246 Free PMC article.
-
Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.Nat Commun. 2024 Oct 7;15(1):8574. doi: 10.1038/s41467-024-52808-2. Nat Commun. 2024. PMID: 39375326 Free PMC article.
-
Cardiovascular adverse effects of antiviral therapies for COVID-19: Evidence and plausible mechanisms.Acta Pharmacol Sin. 2024 Sep 9. doi: 10.1038/s41401-024-01382-w. Online ahead of print. Acta Pharmacol Sin. 2024. PMID: 39251859 Review.
-
Cryo-electron microscopy in the study of virus entry and infection.Front Mol Biosci. 2024 Jul 24;11:1429180. doi: 10.3389/fmolb.2024.1429180. eCollection 2024. Front Mol Biosci. 2024. PMID: 39114367 Free PMC article. Review.
-
Structural determinants of spike infectivity in bat SARS-like coronaviruses RsSHC014 and WIV1.J Virol. 2024 Aug 20;98(8):e0034224. doi: 10.1128/jvi.00342-24. Epub 2024 Jul 19. J Virol. 2024. PMID: 39028202
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

