HSP90 recognizes the N-terminus of huntingtin involved in regulation of huntingtin aggregation by USP19
- PMID: 29093475
- PMCID: PMC5666004
- DOI: 10.1038/s41598-017-13711-7
HSP90 recognizes the N-terminus of huntingtin involved in regulation of huntingtin aggregation by USP19
Abstract
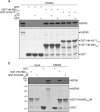
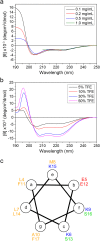
Huntington's disease (HD) is caused by aberrant expansion of polyglutamine (polyQ) in the N-terminus of huntingtin (Htt). Our previous study has demonstrated that HSP90 is involved in the triage decision of Htt, but how HSP90 recognizes and regulates Htt remains elusive. We investigated the interaction between HSP90 and the N-terminal fragments of Htt (Htt-N), such as the N-terminal 90-residue fragment (Htt-N90). Our results showed that HSP90 binds to the N-terminal extreme of Htt-N in a sequence just ahead of the polyQ tract. Structural integration of the middle and C-terminal domains of HSP90 is essential for interacting with Htt-N90, and the dimerization mediated by the C-terminal domain facilitates this interaction. Moreover, ubiquitin-specific protease 19 (USP19), a deubiquitinating enzyme interacting with HSP90, up-regulates the protein level of Htt-N90 and consequently promotes its aggregation, whereas disruption of the interaction between Htt-N90 and HSP90 attenuates the effect of USP19 on Htt-N90. Thus, HSP90 interacts with Htt-N90 on the N-terminal amphipathic α-helix, and then recruits USP19 to modulate the protein level and aggregation of Htt-N90. This study provides mechanistic insights into the recognition between HSP90 and the N-terminus of Htt, and the triage decision for the Htt protein by the HSP90 chaperone system.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Cytoplasmic Ubiquitin-Specific Protease 19 (USP19) Modulates Aggregation of Polyglutamine-Expanded Ataxin-3 and Huntingtin through the HSP90 Chaperone.PLoS One. 2016 Jan 25;11(1):e0147515. doi: 10.1371/journal.pone.0147515. eCollection 2016. PLoS One. 2016. PMID: 26808260 Free PMC article.
-
Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.Biochem J. 2020 Nov 13;477(21):4295-4312. doi: 10.1042/BCJ20200536. Biochem J. 2020. PMID: 33094816
-
Aggregation landscapes of Huntingtin exon 1 protein fragments and the critical repeat length for the onset of Huntington's disease.Proc Natl Acad Sci U S A. 2017 Apr 25;114(17):4406-4411. doi: 10.1073/pnas.1702237114. Epub 2017 Apr 11. Proc Natl Acad Sci U S A. 2017. PMID: 28400517 Free PMC article.
-
Proteostasis of Huntingtin in Health and Disease.Int J Mol Sci. 2017 Jul 19;18(7):1568. doi: 10.3390/ijms18071568. Int J Mol Sci. 2017. PMID: 28753941 Free PMC article. Review.
-
The emerging role of the first 17 amino acids of huntingtin in Huntington's disease.Biomol Concepts. 2015 Mar;6(1):33-46. doi: 10.1515/bmc-2015-0001. Biomol Concepts. 2015. PMID: 25741791 Free PMC article. Review.
Cited by
-
Targeting the proteostasis network in Huntington's disease.Ageing Res Rev. 2019 Jan;49:92-103. doi: 10.1016/j.arr.2018.11.006. Epub 2018 Nov 28. Ageing Res Rev. 2019. PMID: 30502498 Free PMC article. Review.
-
Mechanistic Insights into the Role of Molecular Chaperones in Protein Misfolding Diseases: From Molecular Recognition to Amyloid Disassembly.Int J Mol Sci. 2020 Dec 2;21(23):9186. doi: 10.3390/ijms21239186. Int J Mol Sci. 2020. PMID: 33276458 Free PMC article. Review.
-
Huntingtin Ubiquitination Mechanisms and Novel Possible Therapies to Decrease the Toxic Effects of Mutated Huntingtin.J Pers Med. 2021 Dec 6;11(12):1309. doi: 10.3390/jpm11121309. J Pers Med. 2021. PMID: 34945781 Free PMC article. Review.
-
USP19 deubiquitinates EWS-FLI1 to regulate Ewing sarcoma growth.Sci Rep. 2019 Jan 30;9(1):951. doi: 10.1038/s41598-018-37264-5. Sci Rep. 2019. PMID: 30700749 Free PMC article.
-
HSF1 and Its Role in Huntington's Disease Pathology.Adv Exp Med Biol. 2023;1410:35-95. doi: 10.1007/5584_2022_742. Adv Exp Med Biol. 2023. PMID: 36396925 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

