Repigmentation of Human Vitiligo Skin by NBUVB Is Controlled by Transcription of GLI1 and Activation of the β-Catenin Pathway in the Hair Follicle Bulge Stem Cells
- PMID: 29054607
- PMCID: PMC6135256
- DOI: 10.1016/j.jid.2017.09.040
Repigmentation of Human Vitiligo Skin by NBUVB Is Controlled by Transcription of GLI1 and Activation of the β-Catenin Pathway in the Hair Follicle Bulge Stem Cells
Abstract
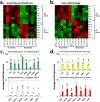
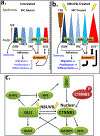
Vitiligo repigmentation is a complex process in which the melanocyte-depleted interfollicular epidermis is repopulated by melanocyte precursors from hair follicle bulge that proliferate, migrate, and differentiate into mature melanocytes on their way to the epidermis. The strongest stimulus for vitiligo repigmentation is narrow-band UVB (NBUVB), but how the hair follicle melanocyte precursors are activated by UV light has not been extensively studied. To better understand this process, we developed an application that combined laser capture microdissection and subsequent whole transcriptome RNA sequencing of hair follicle bulge melanocyte precursors and compared their gene signatures to that of regenerated mature epidermal melanocytes from NBUVB-treated vitiligo skin. Using this strategy, we found up-regulation of TNC, GJB6, and THBS1 in the hair follicle bulge melanocytes and of TYR in the epidermal melanocytes of the NBUVB-treated vitiligo skin. We validated these results by quantitative real-time-PCR using NBUVB-treated vitiligo skin and untreated normal skin. We also identified that GLI1, a candidate stem cell-associated gene, is significantly up-regulated in the melanocytes captured from NBUVB-treated vitiligo bulge compared with untreated vitiligo bulge. These signals are potential key players in the activation of bulge melanocyte precursors during vitiligo repigmentation.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
CONFLICT OF INTEREST
We state no conflict of interest.
Figures


Similar articles
-
Melanocyte Precursors in the Hair Follicle Bulge of Repigmented Vitiligo Skin Are Controlled by RHO-GTPase, KCTD10, and CTNNB1 Signaling.J Invest Dermatol. 2021 Mar;141(3):638-647.e13. doi: 10.1016/j.jid.2020.07.016. Epub 2020 Aug 13. J Invest Dermatol. 2021. PMID: 32800877
-
Isolating RNA from precursor and mature melanocytes from human vitiligo and normal skin using laser capture microdissection.Exp Dermatol. 2016 Oct;25(10):805-11. doi: 10.1111/exd.13072. Exp Dermatol. 2016. PMID: 27193292 Free PMC article.
-
Repigmentation through Melanocyte Regeneration in Vitiligo.Dermatol Clin. 2017 Apr;35(2):205-218. doi: 10.1016/j.det.2016.11.015. Dermatol Clin. 2017. PMID: 28317529 Review.
-
Narrow Band Ultraviolet B Treatment for Human Vitiligo Is Associated with Proliferation, Migration, and Differentiation of Melanocyte Precursors.J Invest Dermatol. 2015 Aug;135(8):2068-2076. doi: 10.1038/jid.2015.126. Epub 2015 Mar 30. J Invest Dermatol. 2015. PMID: 25822579 Free PMC article.
-
Mechanisms of repigmentation induced by photobiomodulation therapy in vitiligo.Exp Dermatol. 2019 Feb;28 Suppl 1:10-14. doi: 10.1111/exd.13823. Exp Dermatol. 2019. PMID: 30698884 Review.
Cited by
-
BMP4-Induced Differentiation of Human Hair Follicle Neural Crest Stem Cells into Precursor Melanocytes from Hair Follicle Bulge.Ann Dermatol. 2020 Oct;32(5):409-416. doi: 10.5021/ad.2020.32.5.409. Epub 2020 Sep 29. Ann Dermatol. 2020. PMID: 33911776 Free PMC article.
-
Effect of Different Wavelengths of Laser Irradiation on the Skin Cells.Int J Mol Sci. 2021 Feb 28;22(5):2437. doi: 10.3390/ijms22052437. Int J Mol Sci. 2021. PMID: 33670977 Free PMC article. Review.
-
Baricitinib is Effective in Treating Progressing Vitiligo in vivo and in vitro.Dose Response. 2022 May 31;20(2):15593258221105370. doi: 10.1177/15593258221105370. eCollection 2022 Apr-Jun. Dose Response. 2022. PMID: 35663493 Free PMC article.
-
A Framework of Major Tumor-Promoting Signal Transduction Pathways Implicated in Melanoma-Fibroblast Dialogue.Cancers (Basel). 2020 Nov 17;12(11):3400. doi: 10.3390/cancers12113400. Cancers (Basel). 2020. PMID: 33212834 Free PMC article. Review.
-
Biological and mechanical influence of three-dimensional microenvironment formed in microwell on multicellular spheroids composed of heterogeneous hair follicle stem cells.Sci Rep. 2023 Dec 20;13(1):22742. doi: 10.1038/s41598-023-49510-6. Sci Rep. 2023. PMID: 38123607 Free PMC article.
References
-
- Amoh Y, Aki R, Hamada Y, Niiyama S, Eshima K, Kawahara K, Sato Y, et al. Nestin-positive hair follicle pluripotent stem cells can promote regeneration of impinged peripheral nerve injury. J Dermatol 2012; 39:33–8. - PubMed
-
- Birlea SA, Goldstein NB, Norris DA. Repigmentation through melanocyte regeneration in vitiligo. Dermatol Clin 2017; 35:205–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

