Analysis of long non-coding RNA expression profiles in ovarian cancer
- PMID: 28789375
- PMCID: PMC5529754
- DOI: 10.3892/ol.2017.6283
Analysis of long non-coding RNA expression profiles in ovarian cancer
Abstract
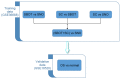
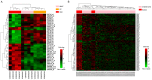
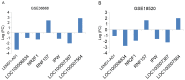
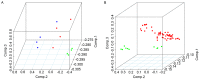
Ovarian cancer is one of the major threats to female health. Identifying cancer cases at an early stage and selecting effective therapeutic drugs for patients is challenging. The number of studies concerning long non-coding RNAs (lncRNAs) is increasing rapidly; there is a large body of evidence indicating that lncRNAs are crucial in oncogenic and tumor-suppression mechanisms. Therefore, in the present study, lncRNA expression in ovarian cancer was considered. All of the existing ovarian cancer microarray datasets in the Gene Expression Omnibus database were assessed and two met the criteria for the present study; these were designated the training and validation sets. A re-annotation pipeline method was established to annotate lncRNAs from existing probe sets. When comparing ovarian cancer with normal ovarian tissues, seven lncRNAs from the RefSeq database, based on their combined ability to classify tissue in the training set, were identified and validated with the validation set. Research into the molecular functions of the seven identified lncRNAs may contribute to the understanding of ovarian cancer oncogenesis; they may also be candidates for novel ovarian cancer biomarkers.
Keywords: long non-coding RNA; microarray; ovarian cancer.
Figures
Similar articles
-
A novel risk score system for assessment of ovarian cancer based on co-expression network analysis and expression level of five lncRNAs.BMC Med Genet. 2019 Jun 10;20(1):103. doi: 10.1186/s12881-019-0832-9. BMC Med Genet. 2019. PMID: 31182053 Free PMC article.
-
Gene microarray analysis of lncRNA and mRNA expression profiles in patients with high‑grade ovarian serous cancer.Int J Mol Med. 2018 Jul;42(1):91-104. doi: 10.3892/ijmm.2018.3588. Epub 2018 Mar 23. Int J Mol Med. 2018. PMID: 29577163 Free PMC article.
-
Microarray expression profiling of long non-coding RNAs in epithelial ovarian cancer.Oncol Lett. 2017 Aug;14(2):2523-2530. doi: 10.3892/ol.2017.6448. Epub 2017 Jun 21. Oncol Lett. 2017. PMID: 28781691 Free PMC article.
-
The role of long non-coding RNA AFAP1-AS1 in human malignant tumors.Pathol Res Pract. 2018 Oct;214(10):1524-1531. doi: 10.1016/j.prp.2018.08.014. Epub 2018 Aug 20. Pathol Res Pract. 2018. PMID: 30173945 Review.
-
Long Non-Coding RNAs: the New Horizon of Gene Regulation in Ovarian Cancer.Cell Physiol Biochem. 2017;44(3):948-966. doi: 10.1159/000485395. Epub 2017 Nov 27. Cell Physiol Biochem. 2017. PMID: 29179183 Review.
Cited by
-
Identification of LncRNA Prognostic Markers for Ovarian Cancer by Integration of Co-expression and CeRNA Network.Front Genet. 2021 Feb 16;11:566497. doi: 10.3389/fgene.2020.566497. eCollection 2020. Front Genet. 2021. PMID: 33664764 Free PMC article.
-
Construction of a lncRNA-miRNA-mRNA network to determine the regulatory roles of lncRNAs in psoriasis.Exp Ther Med. 2019 Nov;18(5):4011-4021. doi: 10.3892/etm.2019.8035. Epub 2019 Sep 23. Exp Ther Med. 2019. PMID: 31611939 Free PMC article.
-
Non-Coding RNAs in Glioma.Cancers (Basel). 2018 Dec 22;11(1):17. doi: 10.3390/cancers11010017. Cancers (Basel). 2018. PMID: 30583549 Free PMC article. Review.
-
Differential protein-coding gene and long noncoding RNA expression in smoking-related lung squamous cell carcinoma.Thorac Cancer. 2017 Nov;8(6):672-681. doi: 10.1111/1759-7714.12510. Epub 2017 Sep 26. Thorac Cancer. 2017. PMID: 28949095 Free PMC article.
-
A Newly Identified lncBCAS1-4_1 Associated With Vitamin D Signaling and EMT in Ovarian Cancer Cells.Front Oncol. 2021 Aug 5;11:691500. doi: 10.3389/fonc.2021.691500. eCollection 2021. Front Oncol. 2021. PMID: 34422647 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
