Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway
- PMID: 28678785
- PMCID: PMC5667900
- DOI: 10.1038/nature23007
Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway
Abstract
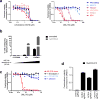
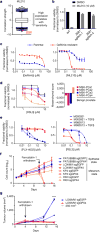
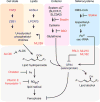
Plasticity of the cell state has been proposed to drive resistance to multiple classes of cancer therapies, thereby limiting their effectiveness. A high-mesenchymal cell state observed in human tumours and cancer cell lines has been associated with resistance to multiple treatment modalities across diverse cancer lineages, but the mechanistic underpinning for this state has remained incompletely understood. Here we molecularly characterize this therapy-resistant high-mesenchymal cell state in human cancer cell lines and organoids and show that it depends on a druggable lipid-peroxidase pathway that protects against ferroptosis, a non-apoptotic form of cell death induced by the build-up of toxic lipid peroxides. We show that this cell state is characterized by activity of enzymes that promote the synthesis of polyunsaturated lipids. These lipids are the substrates for lipid peroxidation by lipoxygenase enzymes. This lipid metabolism creates a dependency on pathways converging on the phospholipid glutathione peroxidase (GPX4), a selenocysteine-containing enzyme that dissipates lipid peroxides and thereby prevents the iron-mediated reactions of peroxides that induce ferroptotic cell death. Dependency on GPX4 was found to exist across diverse therapy-resistant states characterized by high expression of ZEB1, including epithelial-mesenchymal transition in epithelial-derived carcinomas, TGFβ-mediated therapy-resistance in melanoma, treatment-induced neuroendocrine transdifferentiation in prostate cancer, and sarcomas, which are fixed in a mesenchymal state owing to their cells of origin. We identify vulnerability to ferroptic cell death induced by inhibition of a lipid peroxidase pathway as a feature of therapy-resistant cancer cells across diverse mesenchymal cell-state contexts.
Conflict of interest statement
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Figures



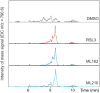

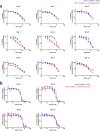
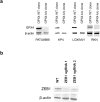

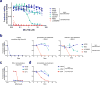


Comment in
-
Therapeutic resistance: Ironing it out.Nat Rev Cancer. 2017 Sep;17(9):510. doi: 10.1038/nrc.2017.71. Epub 2017 Aug 11. Nat Rev Cancer. 2017. PMID: 28798485 No abstract available.
-
Cancer: Ironing it out.Nat Rev Drug Discov. 2017 Sep 1;16(9):602. doi: 10.1038/nrd.2017.168. Nat Rev Drug Discov. 2017. PMID: 28860580 No abstract available.
Similar articles
-
Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis.Proc Natl Acad Sci U S A. 2016 Aug 23;113(34):E4966-75. doi: 10.1073/pnas.1603244113. Epub 2016 Aug 9. Proc Natl Acad Sci U S A. 2016. PMID: 27506793 Free PMC article.
-
Low-density lipoprotein docosahexaenoic acid nanoparticles induce ferroptotic cell death in hepatocellular carcinoma.Free Radic Biol Med. 2017 Nov;112:597-607. doi: 10.1016/j.freeradbiomed.2017.09.002. Epub 2017 Sep 8. Free Radic Biol Med. 2017. PMID: 28893626 Free PMC article.
-
Targeting Dependency on the GPX4 Lipid Peroxide Repair Pathway for Cancer Therapy.Biochemistry. 2018 Apr 10;57(14):2059-2060. doi: 10.1021/acs.biochem.8b00307. Epub 2018 Mar 27. Biochemistry. 2018. PMID: 29584411 Free PMC article. Review. No abstract available.
-
Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition.Nature. 2017 Nov 9;551(7679):247-250. doi: 10.1038/nature24297. Epub 2017 Nov 1. Nature. 2017. PMID: 29088702 Free PMC article.
-
Lipid Peroxidation-Dependent Cell Death Regulated by GPx4 and Ferroptosis.Curr Top Microbiol Immunol. 2017;403:143-170. doi: 10.1007/82_2016_508. Curr Top Microbiol Immunol. 2017. PMID: 28204974 Review.
Cited by
-
Changes in ferrous iron and glutathione promote ferroptosis and frailty in aging Caenorhabditis elegans.Elife. 2020 Jul 21;9:e56580. doi: 10.7554/eLife.56580. Elife. 2020. PMID: 32690135 Free PMC article.
-
Construction and External Validation of a Ferroptosis-Related Gene Signature of Predictive Value for the Overall Survival in Bladder Cancer.Front Mol Biosci. 2021 May 21;8:675651. doi: 10.3389/fmolb.2021.675651. eCollection 2021. Front Mol Biosci. 2021. PMID: 34095228 Free PMC article.
-
Current Progress of Ferroptosis Study in Hepatocellular Carcinoma.Int J Biol Sci. 2024 Jul 1;20(9):3621-3637. doi: 10.7150/ijbs.96014. eCollection 2024. Int J Biol Sci. 2024. PMID: 38993573 Free PMC article. Review.
-
The emerging role of ferroptosis in intestinal disease.Cell Death Dis. 2021 Mar 17;12(4):289. doi: 10.1038/s41419-021-03559-1. Cell Death Dis. 2021. PMID: 33731703 Free PMC article. Review.
-
The Mechanism of Ferroptosis and Applications in Tumor Treatment.Front Pharmacol. 2020 Jul 22;11:1061. doi: 10.3389/fphar.2020.01061. eCollection 2020. Front Pharmacol. 2020. PMID: 32774303 Free PMC article. Review.
References
-
- Hoek KS, et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–656. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM074024/GM/NIGMS NIH HHS/United States
- R35 CA209896/CA/NCI NIH HHS/United States
- U01 CA199253/CA/NCI NIH HHS/United States
- U01 CA176152/CA/NCI NIH HHS/United States
- R01 CA129933/CA/NCI NIH HHS/United States
- R01 CA193837/CA/NCI NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U24 CA194107/CA/NCI NIH HHS/United States
- K08 CA218420/CA/NCI NIH HHS/United States
- P50 CA092629/CA/NCI NIH HHS/United States
- R01 GM038627/GM/NIGMS NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 CA161061/CA/NCI NIH HHS/United States
- R01 GM085081/GM/NIGMS NIH HHS/United States
- U01 CA168397/CA/NCI NIH HHS/United States
- R01 CA097061/CA/NCI NIH HHS/United States
- U01 CA176058/CA/NCI NIH HHS/United States
- R01 CA208100/CA/NCI NIH HHS/United States
- R01 CA154480/CA/NCI NIH HHS/United States
- UL1 TR001102/TR/NCATS NIH HHS/United States
- R01 CA121941/CA/NCI NIH HHS/United States
- U01 CA217848/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

