The emerging field of epigenetics in neurodegeneration and neuroprotection
- PMID: 28515491
- PMCID: PMC6380351
- DOI: 10.1038/nrn.2017.46
The emerging field of epigenetics in neurodegeneration and neuroprotection
Erratum in
-
Author Correction: The emerging field of epigenetics in neurodegeneration and neuroprotection.Nat Rev Neurosci. 2018 Dec;19(12):771. doi: 10.1038/s41583-018-0065-5. Nat Rev Neurosci. 2018. PMID: 30291299
Abstract
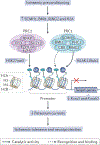
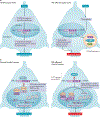
Epigenetic mechanisms - including DNA methylation, histone post-translational modifications and changes in nucleosome positioning - regulate gene expression, cellular differentiation and development in almost all tissues, including the brain. In adulthood, changes in the epigenome are crucial for higher cognitive functions such as learning and memory. Striking new evidence implicates the dysregulation of epigenetic mechanisms in neurodegenerative disorders and diseases. Although these disorders differ in their underlying causes and pathophysiologies, many involve the dysregulation of restrictive element 1-silencing transcription factor (REST), which acts via epigenetic mechanisms to regulate gene expression. Although not somatically heritable, epigenetic modifications in neurons are dynamic and reversible, which makes them good targets for therapeutic intervention.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures
Similar articles
-
Histone Methylation Regulation in Neurodegenerative Disorders.Int J Mol Sci. 2021 Apr 28;22(9):4654. doi: 10.3390/ijms22094654. Int J Mol Sci. 2021. PMID: 33925016 Free PMC article. Review.
-
Epigenetic Modifications in Neurological Diseases: Natural Products as Epigenetic Modulators a Treatment Strategy.Adv Neurobiol. 2016;12:1-25. doi: 10.1007/978-3-319-28383-8_1. Adv Neurobiol. 2016. PMID: 27651245
-
Nutritional influences on epigenetics and age-related disease.Proc Nutr Soc. 2012 Feb;71(1):75-83. doi: 10.1017/S0029665111003302. Epub 2011 Nov 4. Proc Nutr Soc. 2012. PMID: 22051144 Review.
-
Ageing and epigenetics: linking neurodevelopmental and neurodegenerative disorders.Dev Med Child Neurol. 2019 Oct;61(10):1134-1138. doi: 10.1111/dmcn.14210. Epub 2019 Mar 18. Dev Med Child Neurol. 2019. PMID: 30883719 Review.
-
Epigenetics and therapeutic targets mediating neuroprotection.Brain Res. 2015 Dec 2;1628(Pt B):265-272. doi: 10.1016/j.brainres.2015.07.034. Epub 2015 Jul 30. Brain Res. 2015. PMID: 26236020 Free PMC article. Review.
Cited by
-
Epigenetic Changes in Prion and Prion-like Neurodegenerative Diseases: Recent Advances, Potential as Biomarkers, and Future Perspectives.Int J Mol Sci. 2022 Oct 20;23(20):12609. doi: 10.3390/ijms232012609. Int J Mol Sci. 2022. PMID: 36293477 Free PMC article. Review.
-
Epigenetic Regulation of Endothelial Cell Function by Nucleic Acid Methylation in Cardiac Homeostasis and Disease.Cardiovasc Drugs Ther. 2021 Oct;35(5):1025-1044. doi: 10.1007/s10557-020-07019-4. Cardiovasc Drugs Ther. 2021. PMID: 32748033 Free PMC article. Review.
-
Large-scale pathway specific polygenic risk and transcriptomic community network analysis identifies novel functional pathways in Parkinson disease.Acta Neuropathol. 2020 Sep;140(3):341-358. doi: 10.1007/s00401-020-02181-3. Epub 2020 Jun 29. Acta Neuropathol. 2020. PMID: 32601912 Free PMC article.
-
Epigenome-wide meta-analysis of blood DNA methylation and its association with subcortical volumes: findings from the ENIGMA Epigenetics Working Group.Mol Psychiatry. 2021 Aug;26(8):3884-3895. doi: 10.1038/s41380-019-0605-z. Epub 2019 Dec 6. Mol Psychiatry. 2021. PMID: 31811260 Free PMC article.
-
Molecular and epigenetic markers as promising tools to quantify the effect of occupational exposures and the risk of developing non-communicable diseases.Med Lav. 2019 Jun 28;110(3):168-190. doi: 10.23749/mdl.v110i3.8538. Med Lav. 2019. PMID: 31268425 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

