CRISPR/Cas9 Screens Reveal Epstein-Barr Virus-Transformed B Cell Host Dependency Factors
- PMID: 28494239
- PMCID: PMC8938989
- DOI: 10.1016/j.chom.2017.04.005
CRISPR/Cas9 Screens Reveal Epstein-Barr Virus-Transformed B Cell Host Dependency Factors
Abstract
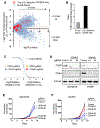
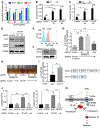
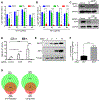
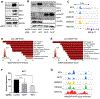
Epstein-Barr virus (EBV) causes endemic Burkitt lymphoma (BL) and immunosuppression-related lymphomas. These B cell malignancies arise by distinct transformation pathways and have divergent viral and host expression programs. To identify host dependency factors resulting from these EBV+, B cell-transformed cell states, we performed parallel genome-wide CRISPR/Cas9 loss-of-function screens in BL and lymphoblastoid cell lines (LCLs). These highlighted 57 BL and 87 LCL genes uniquely important for their growth and survival. LCL hits were enriched for EBV-induced genes, including viral super-enhancer targets. Our systematic approach uncovered key mechanisms by which EBV oncoproteins activate the PI3K/AKT pathway and evade tumor suppressor responses. LMP1-induced cFLIP was found to be critical for LCL defense against TNFα-mediated programmed cell death, whereas EBV-induced BATF/IRF4 were critical for BIM suppression and MYC induction in LCLs. Finally, EBV super-enhancer-targeted IRF2 protected LCLs against Blimp1-mediated tumor suppression. Our results identify viral transformation-driven synthetic lethal targets for therapeutic intervention.
Keywords: CRISPR; Epstein-Barr virus; NF-kappaB; apoptosis; dependency factor; gamma-herpesvirus; interferon regulatory factor; oncoprotein; synthetic lethal; tumor virus.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Epstein-Barr virus nuclear antigen 3A partially coincides with EBNA3C genome-wide and is tethered to DNA through BATF complexes.Proc Natl Acad Sci U S A. 2015 Jan 13;112(2):554-9. doi: 10.1073/pnas.1422580112. Epub 2014 Dec 24. Proc Natl Acad Sci U S A. 2015. PMID: 25540416 Free PMC article.
-
Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-κB activation.J Virol. 2012 Oct;86(20):11096-106. doi: 10.1128/JVI.01069-12. Epub 2012 Aug 1. J Virol. 2012. PMID: 22855490 Free PMC article.
-
RNA Sequencing Analyses of Gene Expression during Epstein-Barr Virus Infection of Primary B Lymphocytes.J Virol. 2019 Jun 14;93(13):e00226-19. doi: 10.1128/JVI.00226-19. Print 2019 Jul 1. J Virol. 2019. PMID: 31019051 Free PMC article.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
-
EBV and Apoptosis: The Viral Master Regulator of Cell Fate?Viruses. 2017 Nov 13;9(11):339. doi: 10.3390/v9110339. Viruses. 2017. PMID: 29137176 Free PMC article. Review.
Cited by
-
Epstein-Barr virus induces germinal center light zone chromatin architecture and promotes survival through enhancer looping at the BCL2A1 locus.mBio. 2024 Jan 16;15(1):e0244423. doi: 10.1128/mbio.02444-23. Epub 2023 Dec 7. mBio. 2024. PMID: 38059622 Free PMC article.
-
BATF regulates the expression of Nfil3, Wnt10a and miR155hg for efficient induction of antibody class switch recombination in mice.Eur J Immunol. 2018 Sep;48(9):1492-1505. doi: 10.1002/eji.201747360. Epub 2018 Jun 26. Eur J Immunol. 2018. PMID: 29898247 Free PMC article.
-
c-Myc Represses Transcription of Epstein-Barr Virus Latent Membrane Protein 1 Early after Primary B Cell Infection.J Virol. 2018 Jan 2;92(2):e01178-17. doi: 10.1128/JVI.01178-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118124 Free PMC article.
-
Epstein-Barr virus lytic gene BNRF1 promotes B-cell lymphomagenesis via IFI27 upregulation.PLoS Pathog. 2024 Feb 1;20(2):e1011954. doi: 10.1371/journal.ppat.1011954. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38300891 Free PMC article.
-
Epigenetic crossroads of the Epstein-Barr virus B-cell relationship.Curr Opin Virol. 2018 Oct;32:15-23. doi: 10.1016/j.coviro.2018.08.012. Epub 2018 Sep 15. Curr Opin Virol. 2018. PMID: 30227386 Free PMC article. Review.
References
-
- Anderton E, Yee J, Smith P, Crook T, White RE, and Allday MJ (2008). Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt’s lymphoma. Oncogene 27, 421–433. - PubMed
-
- Basso K, and Dalla-Favera R (2012). Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev 247, 172–183. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

