The First Report of miRNAs from a Thysanopteran Insect, Thrips palmi Karny Using High-Throughput Sequencing
- PMID: 27685664
- PMCID: PMC5042526
- DOI: 10.1371/journal.pone.0163635
The First Report of miRNAs from a Thysanopteran Insect, Thrips palmi Karny Using High-Throughput Sequencing
Abstract
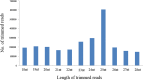
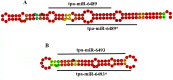
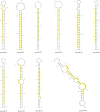
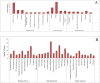
Thrips palmi Karny (Thysanoptera: Thripidae) is the sole vector of Watermelon bud necrosis tospovirus, where the crop loss has been estimated to be around USD 50 million annually. Chemical insecticides are of limited use in the management of T. palmi due to the thigmokinetic behaviour and development of high levels of resistance to insecticides. There is an urgent need to find out an effective futuristic management strategy, where the small RNAs especially microRNAs hold great promise as a key player in the growth and development. miRNAs are a class of short non-coding RNAs involved in regulation of gene expression either by mRNA cleavage or by translational repression. We identified and characterized a total of 77 miRNAs from T. palmi using high-throughput deep sequencing. Functional classifications of the targets for these miRNAs revealed that majority of them are involved in the regulation of transcription and translation, nucleotide binding and signal transduction. We have also validated few of these miRNAs employing stem-loop RT-PCR, qRT-PCR and Northern blot. The present study not only provides an in-depth understanding of the biological and physiological roles of miRNAs in governing gene expression but may also lead as an invaluable tool for the management of thysanopteran insects in the future.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Genome-wide identification, expression profiling, and target gene analysis of microRNAs in the Onion thrips, Thrips tabaci Lindeman (Thysanoptera: Thripidae), vectors of tospoviruses (Bunyaviridae).Ecol Evol. 2018 Jun 7;8(13):6399-6419. doi: 10.1002/ece3.3762. eCollection 2018 Jul. Ecol Evol. 2018. PMID: 30038744 Free PMC article.
-
Molecular differences in the mitochondrial cytochrome oxidase I (mtCOI) gene and development of a species-specific marker for onion thrips, Thrips tabaci Lindeman, and melon thrips, T. palmi Karny (Thysanoptera: Thripidae), vectors of tospoviruses (Bunyaviridae).Bull Entomol Res. 2007 Oct;97(5):461-70. doi: 10.1017/S0007485307005147. Bull Entomol Res. 2007. PMID: 17916265
-
Exposure to watermelon bud necrosis virus and groundnut bud necrosis virus alters the life history traits of their vector, Thrips palmi (Thysanoptera: Thripidae).Arch Virol. 2019 Nov;164(11):2799-2804. doi: 10.1007/s00705-019-04381-z. Epub 2019 Aug 22. Arch Virol. 2019. PMID: 31440810
-
Biotic resistance limits the invasiveness of the western flower thrips, Frankliniella occidentalis (Thysanoptera: Thripidae), in Florida.Insect Sci. 2016 Apr;23(2):175-82. doi: 10.1111/1744-7917.12250. Epub 2015 Sep 23. Insect Sci. 2016. PMID: 26149353 Review.
-
Western flower thrips resistance to insecticides: detection, mechanisms and management strategies.Pest Manag Sci. 2012 Aug;68(8):1111-21. doi: 10.1002/ps.3305. Epub 2012 May 4. Pest Manag Sci. 2012. PMID: 22566175 Review.
Cited by
-
Frontiers Approaches to the Diagnosis of Thrips (Thysanoptera): How Effective Are the Molecular and Electronic Detection Platforms?Insects. 2021 Oct 9;12(10):920. doi: 10.3390/insects12100920. Insects. 2021. PMID: 34680689 Free PMC article. Review.
-
Analysis of Amblyomma americanum microRNAs in response to Ehrlichia chaffeensis infection and their potential role in vectorial capacity.Front Cell Infect Microbiol. 2024 Jul 17;14:1427562. doi: 10.3389/fcimb.2024.1427562. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39086604 Free PMC article.
-
Genome-wide identification, expression profiling, and target gene analysis of microRNAs in the Onion thrips, Thrips tabaci Lindeman (Thysanoptera: Thripidae), vectors of tospoviruses (Bunyaviridae).Ecol Evol. 2018 Jun 7;8(13):6399-6419. doi: 10.1002/ece3.3762. eCollection 2018 Jul. Ecol Evol. 2018. PMID: 30038744 Free PMC article.
-
Analysis of Amblyomma americanum microRNAs in response to Ehrlichia chaffeensis infection and their potential role in vectorial capacity.bioRxiv [Preprint]. 2024 May 6:2024.05.03.592465. doi: 10.1101/2024.05.03.592465. bioRxiv. 2024. Update in: Front Cell Infect Microbiol. 2024 Jul 17;14:1427562. doi: 10.3389/fcimb.2024.1427562. PMID: 38765993 Free PMC article. Updated. Preprint.
-
Transcriptome-wide responses of adult melon thrips (Thrips palmi) associated with capsicum chlorosis virus infection.PLoS One. 2018 Dec 7;13(12):e0208538. doi: 10.1371/journal.pone.0208538. eCollection 2018. PLoS One. 2018. PMID: 30532222 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

