Functional characterization of a monoclonal antibody epitope using a lambda phage display-deep sequencing platform
- PMID: 27530334
- PMCID: PMC4987625
- DOI: 10.1038/srep31458
Functional characterization of a monoclonal antibody epitope using a lambda phage display-deep sequencing platform
Abstract
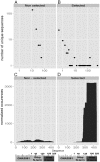
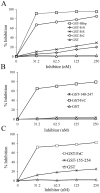
We have recently described a method, named PROFILER, for the identification of antigenic regions preferentially targeted by polyclonal antibody responses after vaccination. To test the ability of the technique to provide insights into the functional properties of monoclonal antibody (mAb) epitopes, we used here a well-characterized epitope of meningococcal factor H binding protein (fHbp), which is recognized by mAb 12C1. An fHbp library, engineered on a lambda phage vector enabling surface expression of polypeptides of widely different length, was subjected to massive parallel sequencing of the phage inserts after affinity selection with the 12C1 mAb. We detected dozens of unique antibody-selected sequences, the most enriched of which (designated as FrC) could largely recapitulate the ability of fHbp to bind mAb 12C1. Computational analysis of the cumulative enrichment of single amino acids in the antibody-selected fragments identified two overrepresented stretches of residues (H248-K254 and S140-G154), whose presence was subsequently found to be required for binding of FrC to mAb 12C1. Collectively, these results suggest that the PROFILER technology can rapidly and reliably identify, in the context of complex conformational epitopes, discrete "hot spots" with a crucial role in antigen-antibody interactions, thereby providing useful clues for the functional characterization of the epitope.
Conflict of interest statement
M.D., V.L.C., C. Be. and G.T. hold shares in Scylla Biotech S.r.l., which has submitted a patent application for the PROFILER technology. E.B and V.M. are full-time employees of GSK Vaccines.
Figures

Similar articles
-
Epitope Mapping of a Monoclonal Antibody Directed against Neisserial Heparin Binding Antigen Using Next Generation Sequencing of Antigen-Specific Libraries.PLoS One. 2016 Aug 10;11(8):e0160702. doi: 10.1371/journal.pone.0160702. eCollection 2016. PLoS One. 2016. PMID: 27508302 Free PMC article.
-
Phage display revisited: Epitope mapping of a monoclonal antibody directed against Neisseria meningitidis adhesin A using the PROFILER technology.MAbs. 2016 May-Jun;8(4):741-50. doi: 10.1080/19420862.2016.1158371. Epub 2016 Mar 10. MAbs. 2016. PMID: 26963435 Free PMC article.
-
Rapid profiling of the antigen regions recognized by serum antibodies using massively parallel sequencing of antigen-specific libraries.PLoS One. 2014 Dec 4;9(12):e114159. doi: 10.1371/journal.pone.0114159. eCollection 2014. PLoS One. 2014. PMID: 25473968 Free PMC article.
-
High throughput functional epitope mapping: revisiting phage display platform to scan target antigen surface.MAbs. 2014;6(6):1368-76. doi: 10.4161/mabs.36144. MAbs. 2014. PMID: 25484050 Free PMC article. Review.
-
Bacteriophage lambda as a vehicle for peptide and protein display.Curr Pharm Biotechnol. 2002 Mar;3(1):23-8. doi: 10.2174/1389201023378481. Curr Pharm Biotechnol. 2002. PMID: 11883504 Review.
Cited by
-
Antibody-Based Protective Immunity against Helminth Infections: Antibody Phage Display Derived Antibodies against BmR1 Antigen.Int J Mol Sci. 2017 Nov 22;18(11):2376. doi: 10.3390/ijms18112376. Int J Mol Sci. 2017. PMID: 29165352 Free PMC article.
-
Structural basis for cooperativity of human monoclonal antibodies to meningococcal factor H-binding protein.Commun Biol. 2019 Jun 26;2:241. doi: 10.1038/s42003-019-0493-4. eCollection 2019. Commun Biol. 2019. PMID: 31263785 Free PMC article.
-
Human protective response induced by meningococcus B vaccine is mediated by the synergy of multiple bactericidal epitopes.Sci Rep. 2018 Feb 27;8(1):3700. doi: 10.1038/s41598-018-22057-7. Sci Rep. 2018. PMID: 29487324 Free PMC article.
-
Cocrystal structure of meningococcal factor H binding protein variant 3 reveals a new crossprotective epitope recognized by human mAb 1E6.FASEB J. 2019 Nov;33(11):12099-12111. doi: 10.1096/fj.201900374R. Epub 2019 Oct 5. FASEB J. 2019. PMID: 31442074 Free PMC article.
-
4CMenB vaccine induces elite cross-protective human antibodies that compete with human factor H for binding to meningococcal fHbp.PLoS Pathog. 2020 Oct 2;16(10):e1008882. doi: 10.1371/journal.ppat.1008882. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33007046 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

