Exome Sequencing Identifies Biallelic MSH3 Germline Mutations as a Recessive Subtype of Colorectal Adenomatous Polyposis
- PMID: 27476653
- PMCID: PMC4974087
- DOI: 10.1016/j.ajhg.2016.06.015
Exome Sequencing Identifies Biallelic MSH3 Germline Mutations as a Recessive Subtype of Colorectal Adenomatous Polyposis
Abstract
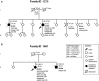
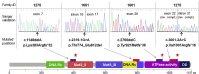
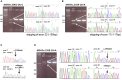
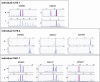
In ∼30% of families affected by colorectal adenomatous polyposis, no germline mutations have been identified in the previously implicated genes APC, MUTYH, POLE, POLD1, and NTHL1, although a hereditary etiology is likely. To uncover further genes with high-penetrance causative mutations, we performed exome sequencing of leukocyte DNA from 102 unrelated individuals with unexplained adenomatous polyposis. We identified two unrelated individuals with differing compound-heterozygous loss-of-function (LoF) germline mutations in the mismatch-repair gene MSH3. The impact of the MSH3 mutations (c.1148delA, c.2319-1G>A, c.2760delC, and c.3001-2A>C) was indicated at the RNA and protein levels. Analysis of the diseased individuals' tumor tissue demonstrated high microsatellite instability of di- and tetranucleotides (EMAST), and immunohistochemical staining illustrated a complete loss of nuclear MSH3 in normal and tumor tissue, confirming the LoF effect and causal relevance of the mutations. The pedigrees, genotypes, and frequency of MSH3 mutations in the general population are consistent with an autosomal-recessive mode of inheritance. Both index persons have an affected sibling carrying the same mutations. The tumor spectrum in these four persons comprised colorectal and duodenal adenomas, colorectal cancer, gastric cancer, and an early-onset astrocytoma. Additionally, we detected one unrelated individual with biallelic PMS2 germline mutations, representing constitutional mismatch-repair deficiency. Potentially causative variants in 14 more candidate genes identified in 26 other individuals require further workup. In the present study, we identified biallelic germline MSH3 mutations in individuals with a suspected hereditary tumor syndrome. Our data suggest that MSH3 mutations represent an additional recessive subtype of colorectal adenomatous polyposis.
Keywords: adenomatous polyposis; candidate genes; exome sequencing; familial colorectal cancer; hereditary tumor syndromes; massive parallel sequencing; mismatch repair.
Copyright © 2016 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Exome sequencing identifies potential novel candidate genes in patients with unexplained colorectal adenomatous polyposis.Fam Cancer. 2016 Apr;15(2):281-8. doi: 10.1007/s10689-016-9870-z. Fam Cancer. 2016. PMID: 26780541
-
Low frequency of AXIN2 mutations and high frequency of MUTYH mutations in patients with multiple polyposis.Hum Mutat. 2006 Oct;27(10):1064. doi: 10.1002/humu.9460. Hum Mutat. 2006. PMID: 16941501
-
Exome sequencing in diagnostic evaluation of colorectal cancer predisposition in young patients.Scand J Gastroenterol. 2013 Jun;48(6):672-8. doi: 10.3109/00365521.2013.783102. Epub 2013 Apr 2. Scand J Gastroenterol. 2013. PMID: 23544471
-
Update on genetic predisposition to colorectal cancer and polyposis.Mol Aspects Med. 2019 Oct;69:10-26. doi: 10.1016/j.mam.2019.03.001. Epub 2019 Mar 18. Mol Aspects Med. 2019. PMID: 30862463 Review.
-
MUTYH-associated colorectal cancer and adenomatous polyposis.Surg Today. 2014 Apr;44(4):593-600. doi: 10.1007/s00595-013-0592-7. Epub 2013 Apr 19. Surg Today. 2014. PMID: 23605219 Review.
Cited by
-
The complementary roles of genome-wide approaches in identifying genes linked to an inherited risk of colorectal cancer.Hered Cancer Clin Pract. 2023 Jan 28;21(1):1. doi: 10.1186/s13053-023-00245-5. Hered Cancer Clin Pract. 2023. PMID: 36707860 Free PMC article. Review.
-
Complex mutation profiles in mismatch repair and ribonucleotide reductase mutants reveal novel repair substrate specificity of MutS homolog (MSH) complexes.Genetics. 2022 Jul 30;221(4):iyac092. doi: 10.1093/genetics/iyac092. Genetics. 2022. PMID: 35686905 Free PMC article.
-
Genotypic and Phenotypic Characteristics of Hereditary Colorectal Cancer.Ann Coloproctol. 2021 Dec;37(6):368-381. doi: 10.3393/ac.2021.00878.0125. Epub 2021 Dec 22. Ann Coloproctol. 2021. PMID: 34961301 Free PMC article. Review.
-
Gene-specific ACMG/AMP classification criteria for germline APC variants: Recommendations from the ClinGen InSiGHT Hereditary Colorectal Cancer/Polyposis Variant Curation Expert Panel.Genet Med. 2024 Feb;26(2):100992. doi: 10.1016/j.gim.2023.100992. Epub 2023 Oct 4. Genet Med. 2024. PMID: 37800450 Free PMC article.
-
Microsatellite Status Detection of Colorectal Cancer: Evaluation of Inconsistency between PCR and IHC.J Cancer. 2023 Apr 17;14(7):1132-1140. doi: 10.7150/jca.81675. eCollection 2023. J Cancer. 2023. PMID: 37215453 Free PMC article.
References
-
- Galiatsatos P., Foulkes W.D. Familial adenomatous polyposis. Am. J. Gastroenterol. 2006;101:385–398. - PubMed
-
- Al-Tassan N., Chmiel N.H., Maynard J., Fleming N., Livingston A.L., Williams G.T., Hodges A.K., Davies D.R., David S.S., Sampson J.R., Cheadle J.P. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat. Genet. 2002;30:227–232. - PubMed
-
- Mazzei F., Viel A., Bignami M. Role of MUTYH in human cancer. Mutat. Res. 2013;743-744:33–43. - PubMed
-
- Krawitz P.M., Schweiger M.R., Rödelsperger C., Marcelis C., Kölsch U., Meisel C., Stephani F., Kinoshita T., Murakami Y., Bauer S. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat. Genet. 2010;42:827–829. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

