Pridopidine activates neuroprotective pathways impaired in Huntington Disease
- PMID: 27466197
- PMCID: PMC5291233
- DOI: 10.1093/hmg/ddw238
Pridopidine activates neuroprotective pathways impaired in Huntington Disease
Abstract
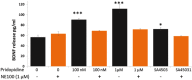
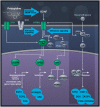
Pridopidine has demonstrated improvement in Huntington Disease (HD) motor symptoms as measured by secondary endpoints in clinical trials. Originally described as a dopamine stabilizer, this mechanism is insufficient to explain the clinical and preclinical effects of pridopidine. This study therefore explored pridopidine's potential mechanisms of action. The effect of pridopidine versus sham treatment on genome-wide expression profiling in the rat striatum was analysed and compared to the pathological expression profile in Q175 knock-in (Q175 KI) vs Q25 WT mouse models. A broad, unbiased pathway analysis was conducted, followed by testing the enrichment of relevant pathways. Pridopidine upregulated the BDNF pathway (P = 1.73E-10), and its effect on BDNF secretion was sigma 1 receptor (S1R) dependent. Many of the same genes were independently found to be downregulated in Q175 KI mice compared to WT (5.2e-7 < P < 0.04). In addition, pridopidine treatment upregulated the glucocorticoid receptor (GR) response, D1R-associated genes and the AKT/PI3K pathway (P = 1E-10, P = 0.001, P = 0.004, respectively). Pridopidine upregulates expression of BDNF, D1R, GR and AKT/PI3K pathways, known to promote neuronal plasticity and survival, as well as reported to demonstrate therapeutic benefit in HD animal models. Activation of S1R, necessary for its effect on the BDNF pathway, represents a core component of the mode of action of pridopidine. Since the newly identified pathways are downregulated in neurodegenerative diseases, including HD, these findings suggest that pridopidine may exert neuroprotective effects beyond its role in alleviating some symptoms of HD.
© The Author 2016. Published by Oxford University Press.
Figures



Similar articles
-
Pridopidine: Overview of Pharmacology and Rationale for its Use in Huntington's Disease.J Huntingtons Dis. 2018;7(1):1-16. doi: 10.3233/JHD-170267. J Huntingtons Dis. 2018. PMID: 29480206 Free PMC article. Review.
-
Large-scale transcriptomic analysis reveals that pridopidine reverses aberrant gene expression and activates neuroprotective pathways in the YAC128 HD mouse.Mol Neurodegener. 2018 May 21;13(1):25. doi: 10.1186/s13024-018-0259-3. Mol Neurodegener. 2018. PMID: 29783994 Free PMC article.
-
The sigma-1 receptor mediates the beneficial effects of pridopidine in a mouse model of Huntington disease.Neurobiol Dis. 2017 Jan;97(Pt A):46-59. doi: 10.1016/j.nbd.2016.10.006. Epub 2016 Nov 3. Neurobiol Dis. 2017. PMID: 27818324 Free PMC article.
-
Pridopidine, a dopamine stabilizer, improves motor performance and shows neuroprotective effects in Huntington disease R6/2 mouse model.J Cell Mol Med. 2015 Nov;19(11):2540-8. doi: 10.1111/jcmm.12604. Epub 2015 Jun 22. J Cell Mol Med. 2015. PMID: 26094900 Free PMC article.
-
Pridopidine for the treatment of Huntington's disease.Expert Opin Investig Drugs. 2016;25(4):485-92. doi: 10.1517/13543784.2016.1153627. Epub 2016 Mar 10. Expert Opin Investig Drugs. 2016. PMID: 26881734 Review.
Cited by
-
Pridopidine: Overview of Pharmacology and Rationale for its Use in Huntington's Disease.J Huntingtons Dis. 2018;7(1):1-16. doi: 10.3233/JHD-170267. J Huntingtons Dis. 2018. PMID: 29480206 Free PMC article. Review.
-
Sigma-1 Receptor (S1R) Interaction with Cholesterol: Mechanisms of S1R Activation and Its Role in Neurodegenerative Diseases.Int J Mol Sci. 2021 Apr 15;22(8):4082. doi: 10.3390/ijms22084082. Int J Mol Sci. 2021. PMID: 33920913 Free PMC article.
-
Current and Possible Future Therapeutic Options for Huntington's Disease.J Cent Nerv Syst Dis. 2022 May 21;14:11795735221092517. doi: 10.1177/11795735221092517. eCollection 2022. J Cent Nerv Syst Dis. 2022. PMID: 35615642 Free PMC article. Review.
-
Early pridopidine treatment improves behavioral and transcriptional deficits in YAC128 Huntington disease mice.JCI Insight. 2017 Dec 7;2(23):e95665. doi: 10.1172/jci.insight.95665. JCI Insight. 2017. PMID: 29212949 Free PMC article.
-
Roles of sigma-1 receptors on mitochondrial functions relevant to neurodegenerative diseases.J Biomed Sci. 2017 Sep 16;24(1):74. doi: 10.1186/s12929-017-0380-6. J Biomed Sci. 2017. PMID: 28917260 Free PMC article. Review.
References
-
- Fisher E.R., Hayden M.R. (2014) Multisource ascertainment of Huntington disease in Canada: Prevalence and population at risk: Multisource Ascertainment of BC HD Patients. Mo.T Disord., 29, 105–114. - PubMed
-
- Dyhring T., Nielsen E., Sonesson C., Pettersson F., Karlsson J., Svensson P., Christophersen P., Waters N. (2010) The dopaminergic stabilizers pridopidine (ACR16) and (−)-OSU6162 display dopamine D2 receptor antagonism and fast receptor dissociation properties. Eur. J. Pharmacol., 628, 19–26. - PubMed
-
- de Yebenes J.G., Landwehrmeyer B., Squitieri F., Reilmann R., Rosser A., Barker R.A., Saft C., Magnet M.K., Sword A., Rembratt Å., et al. (2011) Pridopidine for the treatment of motor function in patients with Huntington’s disease (MermaiHD): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet. Neurol., 10, 1049–1057. - PubMed
-
- Huntington Study Group HART Investigators (2013) A randomized, double-blind, placebo-controlled trial of pridopidine in Huntington’s disease. Mov. Disord., 28, 1407–1415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

