Release of skeletal muscle peptide fragments identifies individual proteins degraded during insulin deprivation in type 1 diabetic humans and mice
- PMID: 27436610
- PMCID: PMC5142007
- DOI: 10.1152/ajpendo.00175.2016
Release of skeletal muscle peptide fragments identifies individual proteins degraded during insulin deprivation in type 1 diabetic humans and mice
Abstract
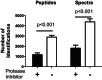
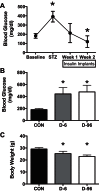
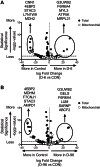
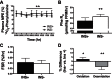
Insulin regulates skeletal muscle protein degradation, but the types of proteins being degraded in vivo remain to be determined due to methodological limitations. We present a method to assess the types of skeletal muscle proteins that are degraded by extracting their degradation products as low-molecular weight (LMW) peptides from muscle samples. High-resolution mass spectrometry was used to identify the original intact proteins that generated the LMW peptides, which we validated in rodents and then applied to humans. We deprived insulin from insulin-treated streptozotocin (STZ) diabetic mice for 6 and 96 h and for 8 h in type 1 diabetic humans (T1D) for comparison with insulin-treated conditions. Protein degradation was measured using activation of autophagy and proteasome pathways, stable isotope tracers, and LMW approaches. In mice, insulin deprivation activated proteasome pathways and autophagy in muscle homogenates and isolated mitochondria. Reproducibility analysis of LMW extracts revealed that ∼80% of proteins were detected consistently. As expected, insulin deprivation increased whole body protein turnover in T1D. Individual protein degradation increased with insulin deprivation, including those involved in mitochondrial function, proteome homeostasis, nDNA support, and contractile/cytoskeleton. Individual mitochondrial proteins that generated more LMW fragment with insulin deprivation included ATP synthase subunit-γ (+0.5-fold, P = 0.007) and cytochrome c oxidase subunit 6 (+0.305-fold, P = 0.03). In conclusion, identifying LMW peptide fragments offers an approach to determine the degradation of individual proteins. Insulin deprivation increases degradation of select proteins and provides insight into the regulatory role of insulin in maintaining proteome homeostasis, especially of mitochondria.
Keywords: autophagy; isotope tracer; low molecular weight; method; peptidomics.
Copyright © 2016 the American Physiological Society.
Figures


Similar articles
-
Altered Skeletal Muscle Mitochondrial Proteome As the Basis of Disruption of Mitochondrial Function in Diabetic Mice.Diabetes. 2016 Mar;65(3):561-73. doi: 10.2337/db15-0823. Epub 2015 Dec 30. Diabetes. 2016. PMID: 26718503 Free PMC article.
-
Effect of insulin deprivation on muscle mitochondrial ATP production and gene transcript levels in type 1 diabetic subjects.Diabetes. 2007 Nov;56(11):2683-9. doi: 10.2337/db07-0378. Epub 2007 Jul 27. Diabetes. 2007. PMID: 17660267
-
Cardiac muscle protein catabolism in diabetes mellitus: activation of the ubiquitin-proteasome system by insulin deficiency.Endocrinology. 2008 Nov;149(11):5384-90. doi: 10.1210/en.2008-0132. Epub 2008 Jul 24. Endocrinology. 2008. PMID: 18653708 Free PMC article.
-
The role of weight loss and exercise in correcting skeletal muscle mitochondrial abnormalities in obesity, diabetes and aging.Mol Cell Endocrinol. 2013 Oct 15;379(1-2):30-4. doi: 10.1016/j.mce.2013.06.018. Epub 2013 Jun 20. Mol Cell Endocrinol. 2013. PMID: 23792186 Review.
-
Recent advances in measuring and understanding the regulation of exercise-mediated protein degradation in skeletal muscle.Am J Physiol Cell Physiol. 2021 Aug 1;321(2):C276-C287. doi: 10.1152/ajpcell.00115.2021. Epub 2021 May 26. Am J Physiol Cell Physiol. 2021. PMID: 34038244 Review.
Cited by
-
Remodeling of skeletal muscle mitochondrial proteome with high-fat diet involves greater changes to β-oxidation than electron transfer proteins in mice.Am J Physiol Endocrinol Metab. 2018 Oct 1;315(4):E425-E434. doi: 10.1152/ajpendo.00051.2018. Epub 2018 May 29. Am J Physiol Endocrinol Metab. 2018. PMID: 29812987 Free PMC article.
-
The role of hormones in sepsis: an integrated overview with a focus on mitochondrial and immune cell dysfunction.Clin Sci (Lond). 2023 May 5;137(9):707-725. doi: 10.1042/CS20220709. Clin Sci (Lond). 2023. PMID: 37144447 Free PMC article. Review.
-
The Effect of Glucagon on Protein Catabolism During Insulin Deficiency: Exchange of Amino Acids Across Skeletal Muscle and the Splanchnic Bed.Diabetes. 2022 Aug 1;71(8):1636-1648. doi: 10.2337/db22-0079. Diabetes. 2022. PMID: 35621914 Free PMC article.
-
Metabolic changes in aging humans: current evidence and therapeutic strategies.J Clin Invest. 2022 Aug 15;132(16):e158451. doi: 10.1172/JCI158451. J Clin Invest. 2022. PMID: 35968789 Free PMC article. Review.
-
Altered mitochondrial function in insulin-deficient and insulin-resistant states.J Clin Invest. 2018 Aug 31;128(9):3671-3681. doi: 10.1172/JCI120843. Epub 2018 Aug 31. J Clin Invest. 2018. PMID: 30168804 Free PMC article. Review.
References
-
- Asmann YW, Stump CS, Short KR, Coenen-Schimke JM, Guo Z, Bigelow ML, Nair KS. Skeletal muscle mitochondrial functions, mitochondrial DNA copy numbers, and gene transcript profiles in type 2 diabetic and nondiabetic subjects at equal levels of low or high insulin and euglycemia. Diabetes 55: 3309–3319, 2006. - PubMed
-
- Biolo G, Fleming RY, Maggi SP, Wolfe RR. Transmembrane transport and intracellular kinetics of amino acids in human skeletal muscle. Am J Physiol Endocrinol Metab 268: E75–E84, 1995. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

