Update on melatonin receptors: IUPHAR Review 20
- PMID: 27314810
- PMCID: PMC4995287
- DOI: 10.1111/bph.13536
Update on melatonin receptors: IUPHAR Review 20
Abstract
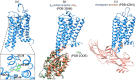
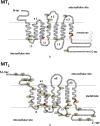
Melatonin receptors are seven transmembrane-spanning proteins belonging to the GPCR superfamily. In mammals, two melatonin receptor subtypes exist - MT1 and MT2 - encoded by the MTNR1A and MTNR1B genes respectively. The current review provides an update on melatonin receptors by the corresponding subcommittee of the International Union of Basic and Clinical Pharmacology. We will highlight recent developments of melatonin receptor ligands, including radioligands, and give an update on the latest phenotyping results of melatonin receptor knockout mice. The current status and perspectives of the structure of melatonin receptor will be summarized. The physiological importance of melatonin receptor dimers and biologically important and type 2 diabetes-associated genetic variants of melatonin receptors will be discussed. The role of melatonin receptors in physiology and disease will be further exemplified by their functions in the immune system and the CNS. Finally, antioxidant and free radical scavenger properties of melatonin and its relation to melatonin receptors will be critically addressed.
© 2016 The British Pharmacological Society.
Figures
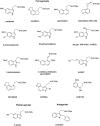
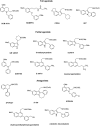
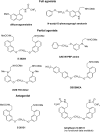
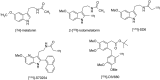
Similar articles
-
Are G protein-coupled receptor heterodimers of physiological relevance?--Focus on melatonin receptors.Chronobiol Int. 2006;23(1-2):419-26. doi: 10.1080/07420520500521863. Chronobiol Int. 2006. PMID: 16687315 Review.
-
Melatonin receptor genes in vertebrates.Int J Mol Sci. 2013 May 27;14(6):11208-23. doi: 10.3390/ijms140611208. Int J Mol Sci. 2013. PMID: 23712359 Free PMC article. Review.
-
Melatonin receptors: molecular pharmacology and signalling in the context of system bias.Br J Pharmacol. 2018 Aug;175(16):3263-3280. doi: 10.1111/bph.13950. Epub 2017 Aug 17. Br J Pharmacol. 2018. PMID: 28707298 Free PMC article. Review.
-
Understanding melatonin receptor pharmacology: latest insights from mouse models, and their relevance to human disease.Bioessays. 2014 Aug;36(8):778-87. doi: 10.1002/bies.201400017. Epub 2014 Jun 5. Bioessays. 2014. PMID: 24903552 Free PMC article. Review.
-
[Sites and mechanisms of action of melatonin in mammals: the MT1 and MT2 receptors].J Soc Biol. 2007;201(1):85-96. doi: 10.1051/jbio:2007010. J Soc Biol. 2007. PMID: 17762828 Review. French.
Cited by
-
Melatonin promotes sleep by activating the BK channel in C. elegans.Proc Natl Acad Sci U S A. 2020 Oct 6;117(40):25128-25137. doi: 10.1073/pnas.2010928117. Epub 2020 Sep 21. Proc Natl Acad Sci U S A. 2020. PMID: 32958651 Free PMC article.
-
The Role of Tryptophan Metabolites in Neuropsychiatric Disorders.Int J Mol Sci. 2022 Sep 1;23(17):9968. doi: 10.3390/ijms23179968. Int J Mol Sci. 2022. PMID: 36077360 Free PMC article. Review.
-
Synthesis, In Silico, and Biological Evaluation of a Borinic Tryptophan-Derivative That Induces Melatonin-like Amelioration of Cognitive Deficit in Male Rat.Int J Mol Sci. 2022 Mar 17;23(6):3229. doi: 10.3390/ijms23063229. Int J Mol Sci. 2022. PMID: 35328650 Free PMC article.
-
Design and Validation of the First Family of Photo-Activatable Ligands for Melatonin Receptors.J Med Chem. 2022 Aug 25;65(16):11229-11240. doi: 10.1021/acs.jmedchem.2c00717. Epub 2022 Aug 5. J Med Chem. 2022. PMID: 35930058 Free PMC article.
-
Sleep in Older Adults and Its Possible Relations With COVID-19.Front Aging Neurosci. 2021 Jun 11;13:647875. doi: 10.3389/fnagi.2021.647875. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34177550 Free PMC article.
References
-
- Adachi A, Natesan AK, Whitfield‐Rucker MG, Weigum SE, Cassone VM (2002). Functional melatonin receptors and metabolic coupling in cultured chick astrocytes. Glia 39: 268–278. - PubMed
-
- Adi N, Mash DC, Ali Y, Singer C, Shehadeh L, Papapetropoulos S (2010). Melatonin MT1 and MT2 receptor expression in Parkinson's disease. Med Sci Monit 16: BR61–BR67. - PubMed
-
- Alarma‐Estrany P, Crooke A, Pintor J (2009). 5‐MCA‐NAT does not act through NQO2 to reduce intraocular pressure in New‐Zealand white rabbit. J Pineal Res 47: 201–209. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

