SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence
- PMID: 27043296
- PMCID: PMC5826646
- DOI: 10.1038/nsmb.3202
SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence
Abstract
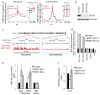
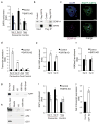
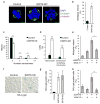
Pericentric heterochromatin silencing at mammalian centromeres is essential for mitotic fidelity and genomic stability. Defective pericentric silencing has been observed in senescent cells, aging tissues, and mammalian tumors, but the underlying mechanisms and functional consequences of these defects are unclear. Here, we uncover an essential role of the human SIRT6 enzyme in pericentric transcriptional silencing, and we show that this function protects against mitotic defects, genomic instability, and cellular senescence. At pericentric heterochromatin, SIRT6 promotes deacetylation of a new substrate, residue K18 of histone H3 (H3K18), and inactivation of SIRT6 in cells leads to H3K18 hyperacetylation and aberrant accumulation of pericentric transcripts. Strikingly, depletion of these transcripts through RNA interference rescues the mitotic and senescence phenotypes of SIRT6-deficient cells. Together, our findings reveal a new function for SIRT6 and regulation of acetylated H3K18 at heterochromatin, and demonstrate the pathogenic role of deregulated pericentric transcription in aging- and cancer-related cellular dysfunction.
Figures

Comment in
-
Chromatin: SIRT6 keeps pericentromeric transcription in check.Nat Rev Mol Cell Biol. 2016 May;17(5):264-5. doi: 10.1038/nrm.2016.46. Epub 2016 Apr 14. Nat Rev Mol Cell Biol. 2016. PMID: 27075411 No abstract available.
Similar articles
-
SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin.Nature. 2008 Mar 27;452(7186):492-6. doi: 10.1038/nature06736. Epub 2008 Mar 12. Nature. 2008. PMID: 18337721 Free PMC article.
-
Chromatin regulation and genome maintenance by mammalian SIRT6.Trends Biochem Sci. 2011 Jan;36(1):39-46. doi: 10.1016/j.tibs.2010.07.009. Epub 2010 Aug 21. Trends Biochem Sci. 2011. PMID: 20729089 Free PMC article. Review.
-
Mammalian SIRT6 Represses Invasive Cancer Cell Phenotypes through ATP Citrate Lyase (ACLY)-Dependent Histone Acetylation.Genes (Basel). 2021 Sep 21;12(9):1460. doi: 10.3390/genes12091460. Genes (Basel). 2021. PMID: 34573442 Free PMC article.
-
SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation.Nature. 2012 Jul 5;487(7405):114-8. doi: 10.1038/nature11043. Nature. 2012. PMID: 22722849 Free PMC article.
-
Sirtuin 6: a review of biological effects and potential therapeutic properties.Mol Biosyst. 2013 Jul;9(7):1789-806. doi: 10.1039/c3mb00001j. Epub 2013 Apr 17. Mol Biosyst. 2013. PMID: 23592245 Review.
Cited by
-
Research advances on silence information regulator 6 as a potential therapeutic target for bone regeneration and repair.Zhejiang Da Xue Xue Bao Yi Xue Ban. 2024 Aug 25;53(4):427-433. doi: 10.3724/zdxbyxb-2023-0615. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 39183069 Free PMC article. Review. Chinese, English.
-
Epigenetic and transcriptional control of adipocyte function by centenarian-associated SIRT6 N308K/A313S mutant.Clin Epigenetics. 2024 Jul 20;16(1):96. doi: 10.1186/s13148-024-01710-1. Clin Epigenetics. 2024. PMID: 39033117 Free PMC article.
-
Oncogenic ETS fusions promote DNA damage and proinflammatory responses via pericentromeric RNAs in extracellular vesicles.J Clin Invest. 2024 Mar 26;134(9):e169470. doi: 10.1172/JCI169470. J Clin Invest. 2024. PMID: 38530366 Free PMC article.
-
Discovery of a potent and highly selective inhibitor of SIRT6 against pancreatic cancer metastasis in vivo.Acta Pharm Sin B. 2024 Mar;14(3):1302-1316. doi: 10.1016/j.apsb.2023.11.014. Epub 2023 Nov 10. Acta Pharm Sin B. 2024. PMID: 38487000 Free PMC article.
-
SIRT3/6: an amazing challenge and opportunity in the fight against fibrosis and aging.Cell Mol Life Sci. 2024 Jan 31;81(1):69. doi: 10.1007/s00018-023-05093-z. Cell Mol Life Sci. 2024. PMID: 38294557 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

