Clip-domain serine proteases as immune factors in insect hemolymph
- PMID: 26688791
- PMCID: PMC4680995
- DOI: 10.1016/j.cois.2015.09.003
Clip-domain serine proteases as immune factors in insect hemolymph
Abstract
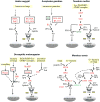
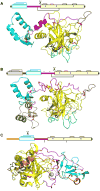
CLIP proteases are non-digestive serine proteases present in hemolymph of insects and other arthropods. They are composed of one or more amino-terminal clip domains followed by a linker sequence and a carboxyl-terminal S1A family serine protease domain. The genes for CLIP proteases have evolved as four clades (CLIPA, CLIPB, CLIPC, CLIPD), each present as multigene families in insect genomes. CLIP proteases in hemolymph function in innate immune responses. These include proteolytic activation of the cytokine Spätzle, to form an active Toll ligand leading to synthesis of antimicrobial peptides, and specific activation of prophenoloxidase, required for the melanization response. CLIP proteases act in cascade pathways. In the immune pathways that have been characterized, microbial surface molecules stimulate activation of an initiating modular serine protease, which then activates a CLIPC, which in turn activates a CLIPB. The active CLIPB then cleaves and activates an effector molecule (proSpätzle or prophenoloxidase). CLIPA proteins are pseudoproteases, lacking proteolytic activity, but some can function as regulators of the activity of other CLIP proteases and form high molecular weight immune complexes. A few three dimensional structures for CLIP proteases are now available for structure-function analysis of these immune factors, revealing structural features that may act in specific activation or in formation of immune complexes. The functions of most CLIP proteases are unknown, even in well studied insect species. It is very likely that additional proteins activated by CLIP proteases and acting in immunity remain to be discovered.
Figures



Similar articles
-
Functional characterization of two clip domain serine proteases in innate immune responses of Aedes aegypti.Parasit Vectors. 2021 Nov 24;14(1):584. doi: 10.1186/s13071-021-05091-9. Parasit Vectors. 2021. PMID: 34819136 Free PMC article.
-
Proteolytic activation and function of the cytokine Spätzle in the innate immune response of a lepidopteran insect, Manduca sexta.FEBS J. 2010 Jan;277(1):148-62. doi: 10.1111/j.1742-4658.2009.07465.x. Epub 2009 Nov 26. FEBS J. 2010. PMID: 19968713 Free PMC article.
-
Hemolymph protease-5 links the melanization and Toll immune pathways in the tobacco hornworm, Manduca sexta.Proc Natl Acad Sci U S A. 2020 Sep 22;117(38):23581-23587. doi: 10.1073/pnas.2004761117. Epub 2020 Sep 8. Proc Natl Acad Sci U S A. 2020. PMID: 32900946 Free PMC article.
-
Serine proteases as mediators of mosquito immune responses.Insect Biochem Mol Biol. 2001 Mar 1;31(3):257-62. doi: 10.1016/s0965-1748(00)00145-4. Insect Biochem Mol Biol. 2001. PMID: 11167095 Review.
-
Drosophila melanogaster clip-domain serine proteases: Structure, function and regulation.Biochimie. 2016 Mar;122:255-69. doi: 10.1016/j.biochi.2015.10.007. Epub 2015 Oct 8. Biochimie. 2016. PMID: 26453810 Review.
Cited by
-
A Plant Virus Ensures Viral Stability in the Hemolymph of Vector Insects through Suppressing Prophenoloxidase Activation.mBio. 2020 Aug 18;11(4):e01453-20. doi: 10.1128/mBio.01453-20. mBio. 2020. PMID: 32817105 Free PMC article.
-
The mosquito melanization response requires hierarchical activation of non-catalytic clip domain serine protease homologs.PLoS Pathog. 2019 Nov 25;15(11):e1008194. doi: 10.1371/journal.ppat.1008194. eCollection 2019 Nov. PLoS Pathog. 2019. PMID: 31765430 Free PMC article.
-
A teratocyte-specific serpin from the endoparasitoid wasp Cotesia vestalis inhibits the prophenoloxidase-activating system of its host Plutella xylostella.Insect Mol Biol. 2022 Apr;31(2):202-215. doi: 10.1111/imb.12751. Epub 2021 Dec 28. Insect Mol Biol. 2022. PMID: 34897868 Free PMC article.
-
Racemization in Post-Translational Modifications Relevance to Protein Aging, Aggregation and Neurodegeneration: Tip of the Iceberg.Symmetry (Basel). 2021 Mar;13(3):455. doi: 10.3390/sym13030455. Epub 2021 Mar 11. Symmetry (Basel). 2021. PMID: 34350031 Free PMC article.
-
The Set of Serine Peptidases of the Tenebrio molitor Beetle: Transcriptomic Analysis on Different Developmental Stages.Int J Mol Sci. 2024 May 25;25(11):5743. doi: 10.3390/ijms25115743. Int J Mol Sci. 2024. PMID: 38891931 Free PMC article.
References
-
- Lange PF, Overall CM. Protein TAILS: when termini tell tales of proteolysis and function. Curr Opin Chem Biol. 2013;17:73–82. - PubMed
-
- Krem MM, Di Cera E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci. 2002;27:67–74. - PubMed
-
- Salvesen GS. New perspectives on proteases. In: Nature Publishing Group, editor. Horizon Symposia. 2004.
-
- Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. - PubMed
-
- Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

