Mechanistic basis of Nek7 activation through Nek9 binding and induced dimerization
- PMID: 26522158
- PMCID: PMC4632185
- DOI: 10.1038/ncomms9771
Mechanistic basis of Nek7 activation through Nek9 binding and induced dimerization
Abstract
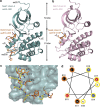
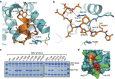
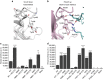
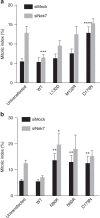
Mitotic spindle assembly requires the regulated activities of protein kinases such as Nek7 and Nek9. Nek7 is autoinhibited by the protrusion of Tyr97 into the active site and activated by the Nek9 non-catalytic C-terminal domain (CTD). CTD binding apparently releases autoinhibition because mutation of Tyr97 to phenylalanine increases Nek7 activity independently of Nek9. Here we find that self-association of the Nek9-CTD is needed for Nek7 activation. We map the minimal Nek7 binding region of Nek9 to residues 810-828. A crystal structure of Nek7(Y97F) bound to Nek9(810-828) reveals a binding site on the C-lobe of the Nek7 kinase domain. Nek7(Y97F) crystallizes as a back-to-back dimer between kinase domain N-lobes, in which the specific contacts within the interface are coupled to the conformation of residue 97. Hence, we propose that the Nek9-CTD activates Nek7 through promoting back-to-back dimerization that releases the autoinhibitory tyrosine residue, a mechanism conserved in unrelated kinase families.
Figures


Similar articles
-
An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek9.Mol Cell. 2009 Nov 25;36(4):560-70. doi: 10.1016/j.molcel.2009.09.038. Mol Cell. 2009. PMID: 19941817 Free PMC article.
-
Structural analysis of the regulation of the DYNLL/LC8 binding to Nek9 by phosphorylation.J Biol Chem. 2013 Apr 26;288(17):12283-94. doi: 10.1074/jbc.M113.459149. Epub 2013 Mar 12. J Biol Chem. 2013. PMID: 23482567 Free PMC article.
-
A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases.J Biol Chem. 2003 Sep 12;278(37):34897-909. doi: 10.1074/jbc.M303663200. Epub 2003 Jul 2. J Biol Chem. 2003. PMID: 12840024
-
DYNLL/LC8 protein controls signal transduction through the Nek9/Nek6 signaling module by regulating Nek6 binding to Nek9.J Biol Chem. 2011 May 20;286(20):18118-29. doi: 10.1074/jbc.M110.209080. Epub 2011 Mar 22. J Biol Chem. 2011. PMID: 21454704 Free PMC article.
-
Characterization of the human NEK7 interactome suggests catalytic and regulatory properties distinct from those of NEK6.J Proteome Res. 2014 Sep 5;13(9):4074-90. doi: 10.1021/pr500437x. Epub 2014 Aug 13. J Proteome Res. 2014. PMID: 25093993 Free PMC article.
Cited by
-
Bruton tyrosine kinase deficiency augments NLRP3 inflammasome activation and causes IL-1β-mediated colitis.J Clin Invest. 2020 Apr 1;130(4):1793-1807. doi: 10.1172/JCI128322. J Clin Invest. 2020. PMID: 31895698 Free PMC article.
-
The established and the predicted roles of dynein light chain in the regulation of mitochondrial apoptosis.Cell Cycle. 2018;17(9):1037-1047. doi: 10.1080/15384101.2018.1464851. Epub 2018 Jul 18. Cell Cycle. 2018. PMID: 30019621 Free PMC article.
-
The Microtubule Regulator NEK7 Coordinates the Wiring of Cortical Parvalbumin Interneurons.Cell Rep. 2018 Jul 31;24(5):1231-1242. doi: 10.1016/j.celrep.2018.06.115. Cell Rep. 2018. PMID: 30067978 Free PMC article.
-
Deep Learning and Structure-Based Virtual Screening for Drug Discovery against NEK7: A Novel Target for the Treatment of Cancer.Molecules. 2022 Jun 25;27(13):4098. doi: 10.3390/molecules27134098. Molecules. 2022. PMID: 35807344 Free PMC article.
-
EML4-ALK V3 oncogenic fusion proteins promote microtubule stabilization and accelerated migration through NEK9 and NEK7.J Cell Sci. 2020 May 11;133(9):jcs241505. doi: 10.1242/jcs.241505. J Cell Sci. 2020. PMID: 32184261 Free PMC article.
References
-
- Endicott J. A., Noble M. E. & Johnson L. N. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 81, 587–613 (2012). - PubMed
-
- Lavoie H., Li J. J., Thevakumaran N., Therrien M. & Sicheri F. Dimerization-induced allostery in protein kinase regulation. Trends Biochem. Sci. 39, 475–486 (2014). - PubMed
-
- Johnson L. N. & Lewis R. J. Structural basis for control by phosphorylation. Chem. Rev. 101, 2209–2242 (2001). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

