Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy
- PMID: 26503055
- PMCID: PMC4779053
- DOI: 10.1038/nature15520
Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy
Abstract
Epigenetic silencing including histone modifications and DNA methylation is an important tumorigenic mechanism. However, its role in cancer immunopathology and immunotherapy is poorly understood. Using human ovarian cancers as our model, here we show that enhancer of zeste homologue 2 (EZH2)-mediated histone H3 lysine 27 trimethylation (H3K27me3) and DNA methyltransferase 1 (DNMT1)-mediated DNA methylation repress the tumour production of T helper 1 (TH1)-type chemokines CXCL9 and CXCL10, and subsequently determine effector T-cell trafficking to the tumour microenvironment. Treatment with epigenetic modulators removes the repression and increases effector T-cell tumour infiltration, slows down tumour progression, and improves the therapeutic efficacy of programmed death-ligand 1 (PD-L1; also known as B7-H1) checkpoint blockade and adoptive T-cell transfusion in tumour-bearing mice. Moreover, tumour EZH2 and DNMT1 are negatively associated with tumour-infiltrating CD8(+) T cells and patient outcome. Thus, epigenetic silencing of TH1-type chemokines is a novel immune-evasion mechanism of tumours. Selective epigenetic reprogramming alters the T-cell landscape in cancer and may enhance the clinical efficacy of cancer therapy.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

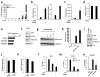
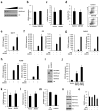

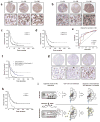
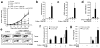
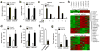
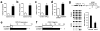

Comment in
-
Tumour immunology: Reducing silence to improve therapy.Nat Rev Immunol. 2015 Dec;15(12):730. doi: 10.1038/nri3941. Epub 2015 Nov 6. Nat Rev Immunol. 2015. PMID: 26542634 No abstract available.
Similar articles
-
PRC2 Epigenetically Silences Th1-Type Chemokines to Suppress Effector T-Cell Trafficking in Colon Cancer.Cancer Res. 2016 Jan 15;76(2):275-82. doi: 10.1158/0008-5472.CAN-15-1938. Epub 2015 Nov 13. Cancer Res. 2016. PMID: 26567139 Free PMC article.
-
DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression.Cancer Lett. 2015 Apr 10;359(2):198-205. doi: 10.1016/j.canlet.2015.01.005. Epub 2015 Jan 13. Cancer Lett. 2015. PMID: 25595591
-
Cooperation between Constitutive and Inducible Chemokines Enables T Cell Engraftment and Immune Attack in Solid Tumors.Cancer Cell. 2019 Jun 10;35(6):885-900.e10. doi: 10.1016/j.ccell.2019.05.004. Cancer Cell. 2019. PMID: 31185212 Free PMC article.
-
Epigenetic strategies synergize with PD-L1/PD-1 targeted cancer immunotherapies to enhance antitumor responses.Acta Pharm Sin B. 2020 May;10(5):723-733. doi: 10.1016/j.apsb.2019.09.006. Epub 2019 Sep 25. Acta Pharm Sin B. 2020. PMID: 32528824 Free PMC article. Review.
-
EZH2 in Bladder Cancer, a Promising Therapeutic Target.Int J Mol Sci. 2015 Nov 13;16(11):27107-32. doi: 10.3390/ijms161126000. Int J Mol Sci. 2015. PMID: 26580594 Free PMC article. Review.
Cited by
-
The Dual Role of STAT1 in Ovarian Cancer: Insight Into Molecular Mechanisms and Application Potentials.Front Cell Dev Biol. 2021 Mar 23;9:636595. doi: 10.3389/fcell.2021.636595. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33834023 Free PMC article. Review.
-
A Randomized Phase II Trial Comparing the Efficacy and Safety of Pioglitazone, Clarithromycin and Metronomic Low-Dose Chemotherapy with Single-Agent Nivolumab Therapy in Patients with Advanced Non-small Cell Lung Cancer Treated in Second or Further Line (ModuLung).Front Pharmacol. 2021 Mar 16;12:599598. doi: 10.3389/fphar.2021.599598. eCollection 2021. Front Pharmacol. 2021. PMID: 33796020 Free PMC article.
-
Molecular subtypes identified by pyroptosis-related genes are associated with tumor microenvironment cell infiltration in colon cancer.Aging (Albany NY). 2022 Nov 16;14(22):9020-9036. doi: 10.18632/aging.204379. Epub 2022 Nov 16. Aging (Albany NY). 2022. PMID: 36384889 Free PMC article.
-
Turning cold tumors into hot tumors by improving T-cell infiltration.Theranostics. 2021 Mar 11;11(11):5365-5386. doi: 10.7150/thno.58390. eCollection 2021. Theranostics. 2021. PMID: 33859752 Free PMC article. Review.
-
PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity.Sci Transl Med. 2020 Jul 8;12(551):eaaz5683. doi: 10.1126/scitranslmed.aaz5683. Sci Transl Med. 2020. PMID: 32641491 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- 5P30CA46592/CA/NCI NIH HHS/United States
- CA099985/CA/NCI NIH HHS/United States
- CA190176/CA/NCI NIH HHS/United States
- CA156685/CA/NCI NIH HHS/United States
- R01 CA193136/CA/NCI NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- R01 GM082856/GM/NIGMS NIH HHS/United States
- F31 CA189440/CA/NCI NIH HHS/United States
- R01 CA171306/CA/NCI NIH HHS/United States
- CA152470/CA/NCI NIH HHS/United States
- CA171306/CA/NCI NIH HHS/United States
- R01 CA152470/CA/NCI NIH HHS/United States
- R01 CA156685/CA/NCI NIH HHS/United States
- R01 CA190176/CA/NCI NIH HHS/United States
- CA123088/CA/NCI NIH HHS/United States
- CA193136/CA/NCI NIH HHS/United States
- R01 CA123088/CA/NCI NIH HHS/United States
- R01 CA099985/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

