Flavivirus Infection Impairs Peroxisome Biogenesis and Early Antiviral Signaling
- PMID: 26423946
- PMCID: PMC4665241
- DOI: 10.1128/JVI.01365-15
Flavivirus Infection Impairs Peroxisome Biogenesis and Early Antiviral Signaling
Abstract
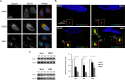
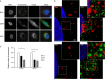
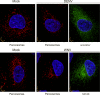
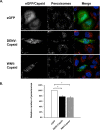
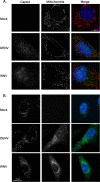
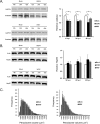
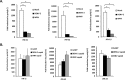
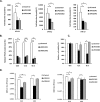
Flaviviruses are significant human pathogens that have an enormous impact on the global health burden. Currently, there are very few vaccines against or therapeutic treatments for flaviviruses, and our understanding of how these viruses cause disease is limited. Evidence suggests that the capsid proteins of flaviviruses play critical nonstructural roles during infection, and therefore, elucidating how these viral proteins affect cellular signaling pathways could lead to novel targets for antiviral therapy. We used affinity purification to identify host cell proteins that interact with the capsid proteins of West Nile and dengue viruses. One of the cellular proteins that formed a stable complex with flavivirus capsid proteins is the peroxisome biogenesis factor Pex19. Intriguingly, flavivirus infection resulted in a significant loss of peroxisomes, an effect that may be due in part to capsid expression. We posited that capsid protein-mediated sequestration and/or degradation of Pex19 results in loss of peroxisomes, a situation that could result in reduced early antiviral signaling. In support of this hypothesis, we observed that induction of the lambda interferon mRNA in response to a viral RNA mimic was reduced by more than 80%. Together, our findings indicate that inhibition of peroxisome biogenesis may be a novel mechanism by which flaviviruses evade the innate immune system during early stages of infection.
Importance: RNA viruses infect hundreds of millions of people each year, causing significant morbidity and mortality. Chief among these pathogens are the flaviviruses, which include dengue virus and West Nile virus. Despite their medical importance, there are very few prophylactic or therapeutic treatments for these viruses. Moreover, the manner in which they subvert the innate immune response in order to establish infection in mammalian cells is not well understood. Recently, peroxisomes were reported to function in early antiviral signaling, but very little is known regarding if or how pathogenic viruses affect these organelles. We report for the first time that flavivirus infection results in significant loss of peroxisomes in mammalian cells, which may indicate that targeting of peroxisomes is a key strategy used by viruses to subvert early antiviral defenses.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Dengue and Zika Virus Capsid Proteins Contain a Common PEX19-Binding Motif.Viruses. 2022 Jan 27;14(2):253. doi: 10.3390/v14020253. Viruses. 2022. PMID: 35215846 Free PMC article.
-
Lipids and flaviviruses, present and future perspectives for the control of dengue, Zika, and West Nile viruses.Prog Lipid Res. 2016 Oct;64:123-137. doi: 10.1016/j.plipres.2016.09.005. Epub 2016 Oct 1. Prog Lipid Res. 2016. PMID: 27702593 Review.
-
STAT5: a Target of Antagonism by Neurotropic Flaviviruses.J Virol. 2019 Nov 13;93(23):e00665-19. doi: 10.1128/JVI.00665-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31534033 Free PMC article.
-
A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection.J Virol. 2017 Mar 29;91(8):e02388-16. doi: 10.1128/JVI.02388-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28148804 Free PMC article.
-
Flavivirus cell entry and membrane fusion.Viruses. 2011 Feb;3(2):160-171. doi: 10.3390/v3020160. Epub 2011 Feb 22. Viruses. 2011. PMID: 22049308 Free PMC article. Review.
Cited by
-
Peroxisomes in host defense.PLoS Pathog. 2020 Jul 2;16(7):e1008636. doi: 10.1371/journal.ppat.1008636. eCollection 2020 Jul. PLoS Pathog. 2020. PMID: 32614930 Free PMC article. No abstract available.
-
Balancing the Opposing Principles That Govern Peroxisome Homeostasis.Trends Biochem Sci. 2021 Mar;46(3):200-212. doi: 10.1016/j.tibs.2020.09.006. Epub 2020 Oct 9. Trends Biochem Sci. 2021. PMID: 33046344 Free PMC article. Review.
-
Peroxisome Plasticity at the Virus-Host Interface.Trends Microbiol. 2019 Nov;27(11):906-914. doi: 10.1016/j.tim.2019.06.006. Epub 2019 Jul 19. Trends Microbiol. 2019. PMID: 31331665 Free PMC article. Review.
-
Dengue and Zika Virus Capsid Proteins Contain a Common PEX19-Binding Motif.Viruses. 2022 Jan 27;14(2):253. doi: 10.3390/v14020253. Viruses. 2022. PMID: 35215846 Free PMC article.
-
The Interplay between Dengue Virus and the Human Innate Immune System: A Game of Hide and Seek.Vaccines (Basel). 2019 Oct 10;7(4):145. doi: 10.3390/vaccines7040145. Vaccines (Basel). 2019. PMID: 31658677 Free PMC article. Review.
References
-
- Pierson TC, Diamond MS. 2013. Flaviviruses, p 747–794. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

