Membrane Curvature Sensing by Amphipathic Helices Is Modulated by the Surrounding Protein Backbone
- PMID: 26366573
- PMCID: PMC4569407
- DOI: 10.1371/journal.pone.0137965
Membrane Curvature Sensing by Amphipathic Helices Is Modulated by the Surrounding Protein Backbone
Abstract
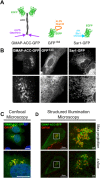
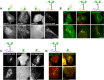
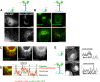
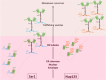
Membrane curvature is involved in numerous biological pathways like vesicle trafficking, endocytosis or nuclear pore complex assembly. In addition to its topological role, membrane curvature is sensed by specific proteins, enabling the coordination of biological processes in space and time. Amongst membrane curvature sensors are the ALPS (Amphipathic Lipid Packing Sensors). ALPS motifs are short peptides with peculiar amphipathic properties. They are found in proteins targeted to distinct curved membranes, mostly in the early secretory pathway. For instance, the ALPS motif of the golgin GMAP210 binds trafficking vesicles, while the ALPS motif of Nup133 targets nuclear pores. It is not clear if, besides curvature sensitivity, ALPS motifs also provide target specificity, or if other domains in the surrounding protein backbone are involved. To elucidate this aspect, we studied the subcellular localization of ALPS motifs outside their natural protein context. The ALPS motifs of GMAP210 or Nup133 were grafted on artificial fluorescent probes. Importantly, ALPS motifs are held in different positions and these contrasting architectures were mimicked by the fluorescent probes. The resulting chimeras recapitulated the original proteins localization, indicating that ALPS motifs are sufficient to specifically localize proteins. Modulating the electrostatic or hydrophobic content of Nup133 ALPS motif modified its avidity for cellular membranes but did not change its organelle targeting properties. In contrast, the structure of the backbone surrounding the helix strongly influenced targeting. In particular, introducing an artificial coiled-coil between ALPS and the fluorescent protein increased membrane curvature sensitivity. This coiled-coil domain also provided membrane curvature sensitivity to the amphipathic helix of Sar1. The degree of curvature sensitivity within the coiled-coil context remains correlated to the natural curvature sensitivity of the helices. This suggests that the chemistry of ALPS motifs is a key parameter for membrane curvature sensitivity, which can be further modulated by the surrounding protein backbone.
Conflict of interest statement
Figures




Similar articles
-
A general amphipathic alpha-helical motif for sensing membrane curvature.Nat Struct Mol Biol. 2007 Feb;14(2):138-46. doi: 10.1038/nsmb1194. Epub 2007 Jan 14. Nat Struct Mol Biol. 2007. PMID: 17220896
-
α-Synuclein and ALPS motifs are membrane curvature sensors whose contrasting chemistry mediates selective vesicle binding.J Cell Biol. 2011 Jul 11;194(1):89-103. doi: 10.1083/jcb.201011118. J Cell Biol. 2011. PMID: 21746853 Free PMC article.
-
The endophilin curvature-sensitive motif requires electrostatic guidance to recycle synaptic vesicles in vivo.Dev Cell. 2022 Mar 28;57(6):750-766.e5. doi: 10.1016/j.devcel.2022.02.021. Epub 2022 Mar 17. Dev Cell. 2022. PMID: 35303431 Free PMC article.
-
Mechanisms of membrane curvature sensing.Annu Rev Biochem. 2011;80:101-23. doi: 10.1146/annurev-biochem-052809-155121. Annu Rev Biochem. 2011. PMID: 21438688 Review.
-
Dunking into the Lipid Bilayer: How Direct Membrane Binding of Nucleoporins Can Contribute to Nuclear Pore Complex Structure and Assembly.Cells. 2021 Dec 20;10(12):3601. doi: 10.3390/cells10123601. Cells. 2021. PMID: 34944108 Free PMC article. Review.
Cited by
-
The N-Terminal Amphipathic Helix of Endophilin Does Not Contribute to Its Molecular Curvature Generation Capacity.J Am Chem Soc. 2016 Nov 9;138(44):14616-14622. doi: 10.1021/jacs.6b06820. Epub 2016 Oct 28. J Am Chem Soc. 2016. PMID: 27755867 Free PMC article.
-
Membrane curvature sensing and symmetry breaking of the M2 proton channel from Influenza A.Elife. 2024 Aug 16;13:e81571. doi: 10.7554/eLife.81571. Elife. 2024. PMID: 39150863 Free PMC article.
-
Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets.Dev Cell. 2018 Jan 8;44(1):73-86.e4. doi: 10.1016/j.devcel.2017.12.011. Epub 2018 Jan 8. Dev Cell. 2018. PMID: 29316443 Free PMC article.
-
Comparative interactomics provides evidence for functional specialization of the nuclear pore complex.Nucleus. 2017 Jul 4;8(4):340-352. doi: 10.1080/19491034.2017.1313936. Epub 2017 May 2. Nucleus. 2017. PMID: 28463551 Free PMC article.
-
A Role for Weak Electrostatic Interactions in Peripheral Membrane Protein Binding.Biophys J. 2016 Mar 29;110(6):1367-78. doi: 10.1016/j.bpj.2016.02.020. Biophys J. 2016. PMID: 27028646 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

