Influenza virus-host interactomes as a basis for antiviral drug development
- PMID: 26364134
- PMCID: PMC5380926
- DOI: 10.1016/j.coviro.2015.08.008
Influenza virus-host interactomes as a basis for antiviral drug development
Abstract
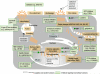
Currently, antiviral drugs that target specific viral protein functions are available for the treatment of influenza; however, concern regarding the emergence of drug-resistant viruses is warranted, as is the urgent need for new antiviral targets, including non-viral targets, such as host cellular factors. Viruses rely on host cellular functions to replicate, and therefore a thorough understanding of the roles of virus-host interactions during influenza virus replication is essential to develop novel anti-influenza drugs that target the host factors involved in virus replication. Here, we review recent studies that used several approaches to identify host factors involved in influenza virus replication. These studies have permitted the construction of an interactome map of virus-host interactions in the influenza virus life cycle, clarifying the entire life cycle of this virus and accelerating the development of new antiviral drugs with a low propensity for the development of resistance.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Disruption of virus-host cell interactions and cell signaling pathways as an anti-viral approach against influenza virus infections.Biol Chem. 2011 Oct;392(10):837-47. doi: 10.1515/BC.2011.121. Biol Chem. 2011. PMID: 21823902 Review.
-
The host interactome of influenza virus presents new potential targets for antiviral drugs.Rev Med Virol. 2011 Nov;21(6):358-69. doi: 10.1002/rmv.703. Epub 2011 Aug 8. Rev Med Virol. 2011. PMID: 21823192 Free PMC article. Review.
-
New-generation screening assays for the detection of anti-influenza compounds targeting viral and host functions.Antiviral Res. 2013 Oct;100(1):120-32. doi: 10.1016/j.antiviral.2013.07.018. Epub 2013 Aug 6. Antiviral Res. 2013. PMID: 23933115 Free PMC article. Review.
-
Network-Guided Discovery of Influenza Virus Replication Host Factors.mBio. 2018 Dec 18;9(6):e02002-18. doi: 10.1128/mBio.02002-18. mBio. 2018. PMID: 30563907 Free PMC article.
-
New drug-strategies to tackle viral-host interactions for the treatment of influenza virus infections.Eur J Pharmacol. 2017 Aug 15;809:178-190. doi: 10.1016/j.ejphar.2017.05.038. Epub 2017 May 19. Eur J Pharmacol. 2017. PMID: 28533172 Review.
Cited by
-
Potential anti-influenza effective plants used in Turkish folk medicine: A review.J Ethnopharmacol. 2021 Jan 30;265:113319. doi: 10.1016/j.jep.2020.113319. Epub 2020 Aug 31. J Ethnopharmacol. 2021. PMID: 32882361 Free PMC article. Review.
-
Inhibition of RAN attenuates influenza a virus replication and nucleoprotein nuclear export.Emerg Microbes Infect. 2024 Dec;13(1):2387910. doi: 10.1080/22221751.2024.2387910. Epub 2024 Aug 12. Emerg Microbes Infect. 2024. PMID: 39087696 Free PMC article.
-
The Influenza Virus Polymerase Complex: An Update on Its Structure, Functions, and Significance for Antiviral Drug Design.Med Res Rev. 2016 Nov;36(6):1127-1173. doi: 10.1002/med.21401. Epub 2016 Aug 29. Med Res Rev. 2016. PMID: 27569399 Free PMC article. Review.
-
Cellular DNA Topoisomerases Are Required for the Synthesis of Hepatitis B Virus Covalently Closed Circular DNA.J Virol. 2019 May 15;93(11):e02230-18. doi: 10.1128/JVI.02230-18. Print 2019 Jun 1. J Virol. 2019. PMID: 30867306 Free PMC article.
-
Non-human primate orthologues of TMPRSS2 cleave and activate the influenza virus hemagglutinin.PLoS One. 2017 May 11;12(5):e0176597. doi: 10.1371/journal.pone.0176597. eCollection 2017. PLoS One. 2017. PMID: 28493964 Free PMC article.
References
-
- Wright PF, Neumann G, Kawaoka Y. In: Orthomyxoviruses. Fields Virology. 6 th edition Knipe PMH DM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 1186–1243.
-
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. - PubMed
-
- Webster RG, Govorkova EA. H5N1 influenza--continuing evolution and spread. N Engl J Med. 2006;355:2174–2177. - PubMed
-
- Yen HL, Webster RG. Pandemic influenza as a current threat. Curr Top Microbiol Immunol. 2009;333:3–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

