Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein
- PMID: 26320998
- PMCID: PMC4567405
- DOI: 10.1016/j.chom.2015.07.015
Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein
Abstract
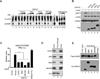
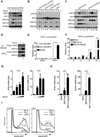
Invading viral DNA can be recognized by the host cytosolic DNA sensor, cyclic GMP-AMP (cGAMP) synthase (cGAS), resulting in production of the second messenger cGAMP, which directs the adaptor protein STING to stimulate production of type I interferons (IFNs). Although several DNA viruses are sensed by cGAS, viral strategies targeting cGAS are virtually unknown. We report here that Kaposi's sarcoma-associated herpesvirus (KSHV) ORF52, an abundant gammaherpesvirus-specific tegument protein, subverts cytosolic DNA sensing by directly inhibiting cGAS enzymatic activity through a mechanism involving both cGAS binding and DNA binding. Moreover, ORF52 homologs in other gammaherpesviruses also inhibit cGAS activity and similarly bind cGAS and DNA, suggesting conserved inhibitory mechanisms. Furthermore, KSHV infection evokes cGAS-dependent responses that can limit the infection, and an ORF52 null mutant exhibits increased cGAS signaling. Our findings reveal a mechanism through which gammaherpesviruses antagonize host cGAS DNA sensing.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
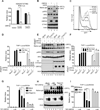
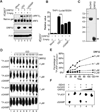
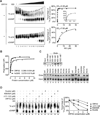
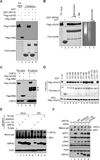
Comment in
-
Blowing Off Steam: Virus Inhibition of cGAS DNA Sensing.Cell Host Microbe. 2015 Sep 9;18(3):270-2. doi: 10.1016/j.chom.2015.08.012. Cell Host Microbe. 2015. PMID: 26355212 Free PMC article.
Similar articles
-
Kaposi's Sarcoma-Associated Herpesvirus Inhibitor of cGAS (KicGAS), Encoded by ORF52, Is an Abundant Tegument Protein and Is Required for Production of Infectious Progeny Viruses.J Virol. 2016 May 12;90(11):5329-5342. doi: 10.1128/JVI.02675-15. Print 2016 Jun 1. J Virol. 2016. PMID: 27009954 Free PMC article.
-
Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses.Proc Natl Acad Sci U S A. 2015 Aug 4;112(31):E4306-15. doi: 10.1073/pnas.1503831112. Epub 2015 Jul 21. Proc Natl Acad Sci U S A. 2015. PMID: 26199418 Free PMC article.
-
Herpes Simplex Virus 1 Abrogates the cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway via Its Virion Host Shutoff Protein, UL41.J Virol. 2017 Feb 28;91(6):e02414-16. doi: 10.1128/JVI.02414-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077645 Free PMC article.
-
KSHV strategies for host dsDNA sensing machinery.Virol Sin. 2016 Dec;31(6):466-471. doi: 10.1007/s12250-016-3877-3. Epub 2016 Dec 5. Virol Sin. 2016. PMID: 27933565 Free PMC article. Review.
-
Conserved strategies for pathogen evasion of cGAS-STING immunity.Curr Opin Immunol. 2020 Oct;66:27-34. doi: 10.1016/j.coi.2020.04.002. Epub 2020 Apr 15. Curr Opin Immunol. 2020. PMID: 32339908 Free PMC article. Review.
Cited by
-
Cyclic Guanosine Monophosphate-Adenosine Monophosphate Synthase (cGAS), a Multifaceted Platform of Intracellular DNA Sensing.Front Immunol. 2021 Feb 23;12:637399. doi: 10.3389/fimmu.2021.637399. eCollection 2021. Front Immunol. 2021. PMID: 33708225 Free PMC article. Review.
-
Myxoma virus lacking the host range determinant M062 stimulates cGAS-dependent type 1 interferon response and unique transcriptomic changes in human monocytes/macrophages.PLoS Pathog. 2022 Sep 14;18(9):e1010316. doi: 10.1371/journal.ppat.1010316. eCollection 2022 Sep. PLoS Pathog. 2022. PMID: 36103568 Free PMC article.
-
Epstein-Barr Viruses: Their Immune Evasion Strategies and Implications for Autoimmune Diseases.Int J Mol Sci. 2024 Jul 26;25(15):8160. doi: 10.3390/ijms25158160. Int J Mol Sci. 2024. PMID: 39125729 Free PMC article. Review.
-
Isolation and Characterization of a Novel Gammaherpesvirus from a Microbat Cell Line.mSphere. 2016 Feb 17;1(1):e00070-15. doi: 10.1128/mSphere.00070-15. eCollection 2016 Jan-Feb. mSphere. 2016. PMID: 27303702 Free PMC article.
-
A Tug of War: DNA-Sensing Antiviral Innate Immunity and Herpes Simplex Virus Type I Infection.Front Microbiol. 2019 Nov 26;10:2627. doi: 10.3389/fmicb.2019.02627. eCollection 2019. Front Microbiol. 2019. PMID: 31849849 Free PMC article. Review.
References
-
- Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, Hornung V. TREX1 Deficiency Triggers Cell-Autonomous Immunity in a cGAS-Dependent Manner. J Immunol. 2014;192:5993–5997. - PubMed
-
- Ahn J, Barber GN. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr Opin Immunol. 2014;31C:121–126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

