CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA
- PMID: 26272696
- PMCID: PMC4536801
- DOI: 10.1186/s12943-015-0426-x
CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA
Abstract
Background: CD90+ liver cancer cells have been described as cancer stem-cell-like (CSC), displaying aggressive and metastatic phenotype. Using two different in vitro models, already described as CD90+ liver cancer stem cells, our aim was to study their interaction with endothelial cells mediated by the release of exosomes.
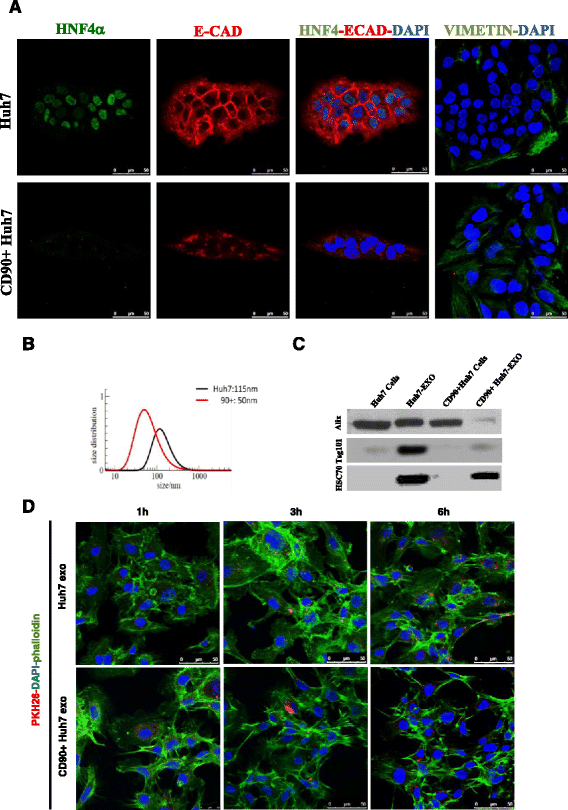
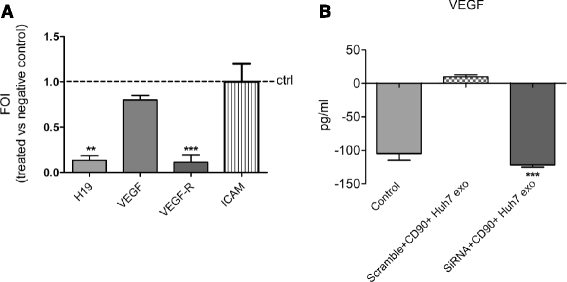
Methods: Exosomes were isolated and characterized from both liver CD90+ cells and hepatoma cell lines. Endothelial cells were treated with exosomes, as well as transfected with a plasmid containing the full length sequence of the long non-coding RNA (lncRNA) H19. Molecular and functional analyses were done to characterize the endothelial phenotype after treatments.
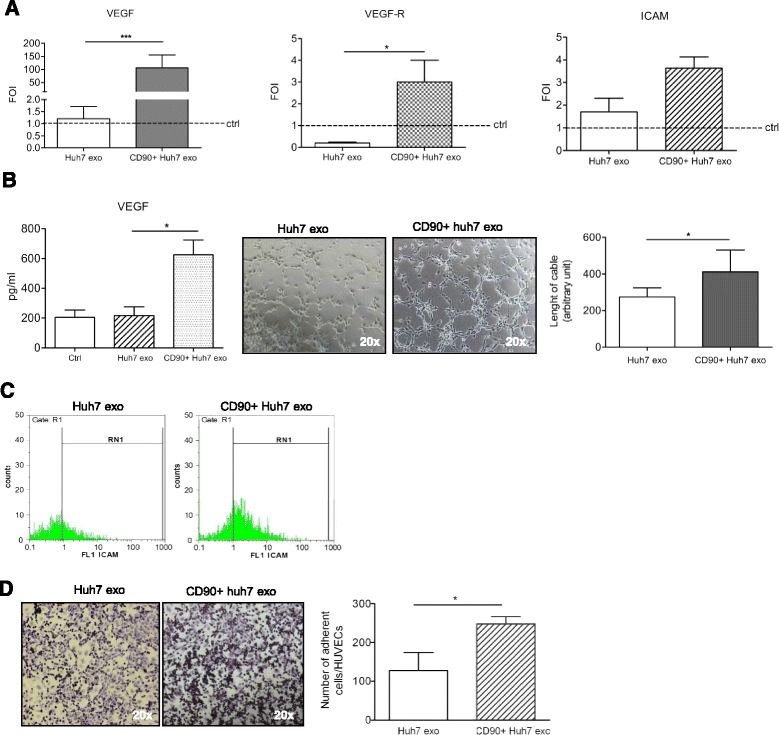
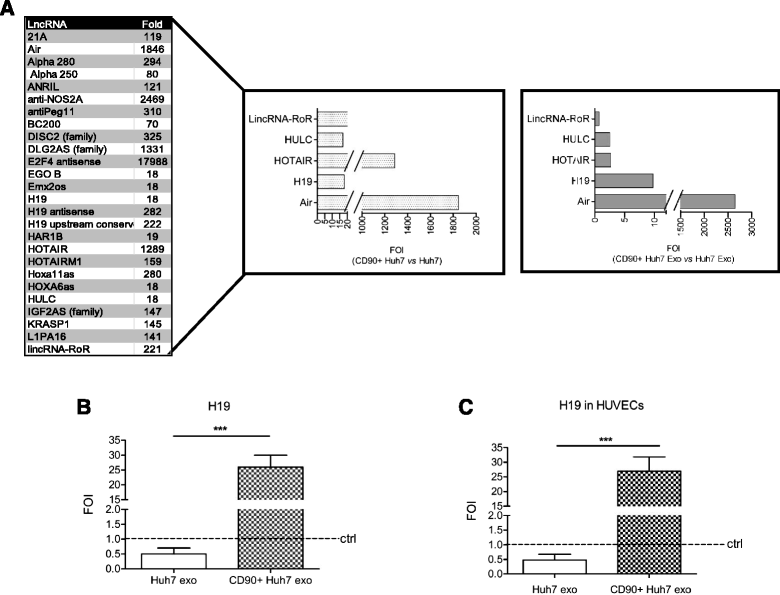
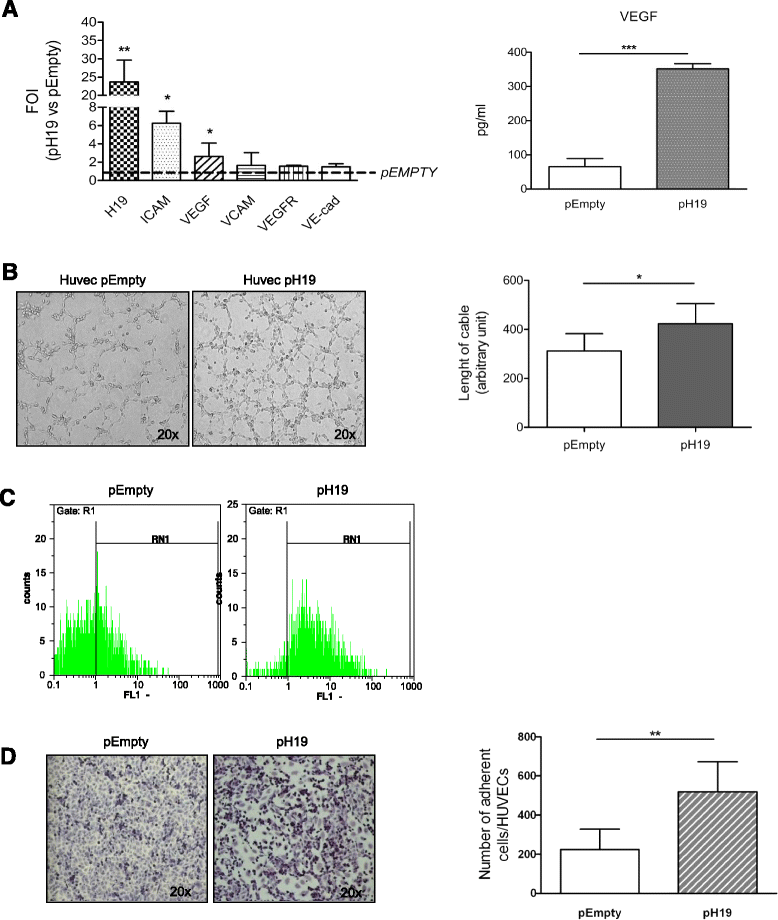
Results: Exosomes released by CD90+ cancer cells, but not by parental hepatoma cells, modulated endothelial cells, promoting angiogenic phenotype and cell-to-cell adhesion. LncRNA profiling revealed that CD90+ cells were enriched in lncRNA H19, and released this through exosomes. Experiments of gain and loss of function of H19 showed that this LncRNA plays an important role in the exosome-mediated phenotype of endothelial cells.
Conclusions: Our data indicate a new exosome-mediated mechanism by which CSC-like CD90+ cells could influence their tumor microenvironment by promoting angiogenesis. Moreover, we suggest the lncRNA H19 as a putative therapeutic target in hepatocellular carcinoma.
Figures
Similar articles
-
Tumor‑released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non‑small cell lung cancer.Oncol Rep. 2018 Dec;40(6):3438-3446. doi: 10.3892/or.2018.6762. Epub 2018 Oct 3. Oncol Rep. 2018. PMID: 30542738 Free PMC article.
-
Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells.J Neurosurg. 2016 Jan;124(1):129-36. doi: 10.3171/2014.12.JNS1426. Epub 2015 Aug 14. J Neurosurg. 2016. PMID: 26274999
-
Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2.Oncol Rep. 2017 Aug;38(2):785-798. doi: 10.3892/or.2017.5742. Epub 2017 Jun 22. Oncol Rep. 2017. PMID: 28656228 Free PMC article.
-
[Advances in the role of exosome-derived long non-coding RNAs in hepatocellular carcinoma].Zhonghua Gan Zang Bing Za Zhi. 2019 Jan 20;27(1):77-81. doi: 10.3760/cma.j.issn.1007-3418.2019.01.019. Zhonghua Gan Zang Bing Za Zhi. 2019. PMID: 30685932 Review. Chinese.
-
Multiple roles of CD90 in cancer.Tumour Biol. 2016 Sep;37(9):11611-11622. doi: 10.1007/s13277-016-5112-0. Epub 2016 Jun 23. Tumour Biol. 2016. PMID: 27337957 Review.
Cited by
-
LncRNA LINC01305 promotes cervical cancer progression through KHSRP and exosome-mediated transfer.Aging (Albany NY). 2021 Feb 26;13(15):19230-19242. doi: 10.18632/aging.202565. Epub 2021 Feb 26. Aging (Albany NY). 2021. PMID: 33638945 Free PMC article.
-
Tumor cells derived-exosomes as angiogenenic agents: possible therapeutic implications.J Transl Med. 2020 Jun 22;18(1):249. doi: 10.1186/s12967-020-02426-5. J Transl Med. 2020. PMID: 32571337 Free PMC article. Review.
-
PDPN+ CAFs facilitate the motility of OSCC cells by inhibiting ferroptosis via transferring exosomal lncRNA FTX.Cell Death Dis. 2023 Nov 22;14(11):759. doi: 10.1038/s41419-023-06280-3. Cell Death Dis. 2023. PMID: 37993428 Free PMC article.
-
Mechanisms of drug resistance in breast cancer liver metastases: Dilemmas and opportunities.Mol Ther Oncolytics. 2023 Feb 6;28:212-229. doi: 10.1016/j.omto.2023.02.001. eCollection 2023 Mar 16. Mol Ther Oncolytics. 2023. PMID: 36860815 Free PMC article. Review.
-
Molecular functions and therapeutic applications of exosomal noncoding RNAs in cancer.Exp Mol Med. 2022 Mar;54(3):216-225. doi: 10.1038/s12276-022-00744-w. Epub 2022 Mar 29. Exp Mol Med. 2022. PMID: 35352001 Free PMC article. Review.
References
-
- Yoo DJ, Kim KM, Jin YJ, Shim JH, Ko GY, Yoon HK, et al. Clinical outcome of 251 patients with extrahepatic metastasis at initial diagnosis of hepatocellular carcinoma: does transarterial chemoembolization improve survival in these patients? J Gastroenterol Hepatol. 2011;26(1):145–54. doi: 10.1111/j.1440-1746.2010.06341.x. - DOI - PubMed
-
- Roncalli M, Park YN, Di Tommaso L. Histopathological classification of hepatocellular carcinoma. Digestive and liver disease. Off J Italian Soc Gastroenterol Italian Assoc Study Liver. 2010;42(Suppl 3):S228–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

