Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination
- PMID: 26269185
- PMCID: PMC4580176
- DOI: 10.1128/JVI.01048-15
Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination
Abstract
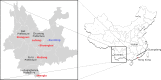
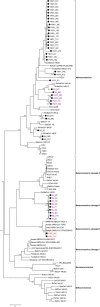
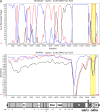
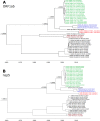
Despite the identification of horseshoe bats as the reservoir of severe acute respiratory syndrome (SARS)-related coronaviruses (SARSr-CoVs), the origin of SARS-CoV ORF8, which contains the 29-nucleotide signature deletion among human strains, remains obscure. Although two SARS-related Rhinolophus sinicus bat CoVs (SARSr-Rs-BatCoVs) previously detected in Chinese horseshoe bats (Rhinolophus sinicus) in Yunnan, RsSHC014 and Rs3367, possessed 95% genome identities to human and civet SARSr-CoVs, their ORF8 protein exhibited only 32.2 to 33% amino acid identities to that of human/civet SARSr-CoVs. To elucidate the origin of SARS-CoV ORF8, we sampled 348 bats of various species in Yunnan, among which diverse alphacoronaviruses and betacoronaviruses, including potentially novel CoVs, were identified, with some showing potential interspecies transmission. The genomes of two betacoronaviruses, SARSr-Rf-BatCoV YNLF_31C and YNLF_34C, from greater horseshoe bats (Rhinolophus ferrumequinum), possessed 93% nucleotide identities to human/civet SARSr-CoV genomes. Although these two betacoronaviruses displayed lower similarities than SARSr-Rs-BatCoV RsSHC014 and Rs3367 in S protein to civet SARSr-CoVs, their ORF8 proteins demonstrated exceptionally high (80.4 to 81.3%) amino acid identities to that of human/civet SARSr-CoVs, compared to SARSr-BatCoVs from other horseshoe bats (23.2 to 37.3%). Potential recombination events were identified around ORF8 between SARSr-Rf-BatCoVs and SARSr-Rs-BatCoVs, leading to the generation of civet SARSr-CoVs. The expression of ORF8 subgenomic mRNA suggested that the ORF8 protein may be functional in SARSr-Rf-BatCoVs. The high Ka/Ks ratio among human SARS-CoVs compared to that among SARSr-BatCoVs supported that ORF8 is under strong positive selection during animal-to-human transmission. Molecular clock analysis using ORF1ab showed that SARSr-Rf-BatCoV YNLF_31C and YNLF_34C diverged from civet/human SARSr-CoVs in approximately 1990. SARS-CoV ORF8 originated from SARSr-CoVs of greater horseshoe bats through recombination, which may be important for animal-to-human transmission.
Importance: Although horseshoe bats are the primary reservoir of SARS-related coronaviruses (SARSr-CoVs), it is still unclear how these bat viruses have evolved to cross the species barrier to infect civets and humans. Most human SARS-CoV epidemic strains contain a signature 29-nucleotide deletion in ORF8, compared to civet SARSr-CoVs, suggesting that ORF8 may be important for interspecies transmission. However, the origin of SARS-CoV ORF8 remains obscure. In particular, SARSr-Rs-BatCoVs from Chinese horseshoe bats (Rhinolophus sinicus) exhibited <40% amino acid identities to human/civet SARS-CoV in the ORF8 protein. We detected diverse alphacoronaviruses and betacoronaviruses among various bat species in Yunnan, China, including two SARSr-Rf-BatCoVs from greater horseshoe bats that possessed ORF8 proteins with exceptionally high amino acid identities to that of human/civet SARSr-CoVs. We demonstrated recombination events around ORF8 between SARSr-Rf-BatCoVs and SARSr-Rs-BatCoVs, leading to the generation of civet SARSr-CoVs. Our findings offer insight into the evolutionary origin of SARS-CoV ORF8 protein, which was likely acquired from SARSr-CoVs of greater horseshoe bats through recombination.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events.J Virol. 2010 Mar;84(6):2808-19. doi: 10.1128/JVI.02219-09. Epub 2010 Jan 13. J Virol. 2010. PMID: 20071579 Free PMC article.
-
Molecular epidemiology, evolution and phylogeny of SARS coronavirus.Infect Genet Evol. 2019 Jul;71:21-30. doi: 10.1016/j.meegid.2019.03.001. Epub 2019 Mar 4. Infect Genet Evol. 2019. PMID: 30844511 Free PMC article. Review.
-
Epidemiology and Genomic Characterization of Two Novel SARS-Related Coronaviruses in Horseshoe Bats from Guangdong, China.mBio. 2022 Jun 28;13(3):e0046322. doi: 10.1128/mbio.00463-22. Epub 2022 Apr 25. mBio. 2022. PMID: 35467426 Free PMC article.
-
Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus.PLoS Pathog. 2017 Nov 30;13(11):e1006698. doi: 10.1371/journal.ppat.1006698. eCollection 2017 Nov. PLoS Pathog. 2017. PMID: 29190287 Free PMC article.
-
Interspecies Jumping of Bat Coronaviruses.Viruses. 2021 Oct 29;13(11):2188. doi: 10.3390/v13112188. Viruses. 2021. PMID: 34834994 Free PMC article. Review.
Cited by
-
Emergence of crucial evidence catalyzing the origin tracing of SARS-CoV-2.PLoS One. 2024 Aug 30;19(8):e0309557. doi: 10.1371/journal.pone.0309557. eCollection 2024. PLoS One. 2024. PMID: 39213297 Free PMC article.
-
Genetic drift promotes and recombination hinders speciation on holey fitness landscapes.PLoS Genet. 2024 Jan 22;20(1):e1011126. doi: 10.1371/journal.pgen.1011126. eCollection 2024 Jan. PLoS Genet. 2024. PMID: 38252672 Free PMC article.
-
Detection of SARS-CoV-2 Δ426 ORF8 Deletion Mutant Cluster in NGS Screening.Microorganisms. 2023 Sep 23;11(10):2378. doi: 10.3390/microorganisms11102378. Microorganisms. 2023. PMID: 37894036 Free PMC article.
-
A comprehensive dataset of animal-associated sarbecoviruses.Sci Data. 2023 Oct 7;10(1):681. doi: 10.1038/s41597-023-02558-5. Sci Data. 2023. PMID: 37805633 Free PMC article.
-
Potential Therapeutic Target and Vaccines for SARS-CoV-2.Pathogens. 2023 Jul 10;12(7):926. doi: 10.3390/pathogens12070926. Pathogens. 2023. PMID: 37513773 Free PMC article.
References
-
- de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya A, Holmes KV, Perlman S, Poon L, Rottier PJ, Talbot PJ, Woo PC, Ziebuhr J. 2011. Coronaviridae, p 806–828. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy, classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses, International Union of Microbiological Societies, Virology Division. Elsevier Academic Press, San Diego, CA.
-
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. 2012. Discovery of seven novel mammalian and avian coronaviruses in Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J Virol 86:3995–4008. doi:10.1128/JVI.06540-11. - DOI - PMC - PubMed
-
- Woo PC, Wang M, Lau SK, Xu H, Poon RW, Guo R, Wong BH, Gao K, Tsoi HW, Huang Y, Li KS, Lam CS, Chan KH, Zheng BJ, Yuen KY. 2007. Comparative analysis of 12 genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol 81:1574–1585. doi:10.1128/JVI.02182-06. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

