Bipartite structure of the inactive mouse X chromosome
- PMID: 26248554
- PMCID: PMC4539712
- DOI: 10.1186/s13059-015-0728-8
Bipartite structure of the inactive mouse X chromosome
Abstract
Background: In mammals, one of the female X chromosomes and all imprinted genes are expressed exclusively from a single allele in somatic cells. To evaluate structural changes associated with allelic silencing, we have applied a recently developed Hi-C assay that uses DNase I for chromatin fragmentation to mouse F1 hybrid systems.
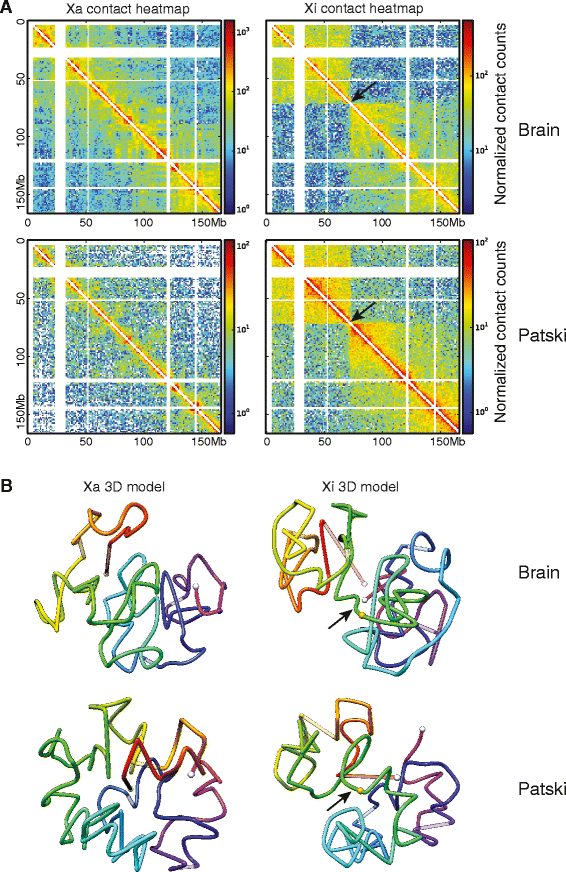
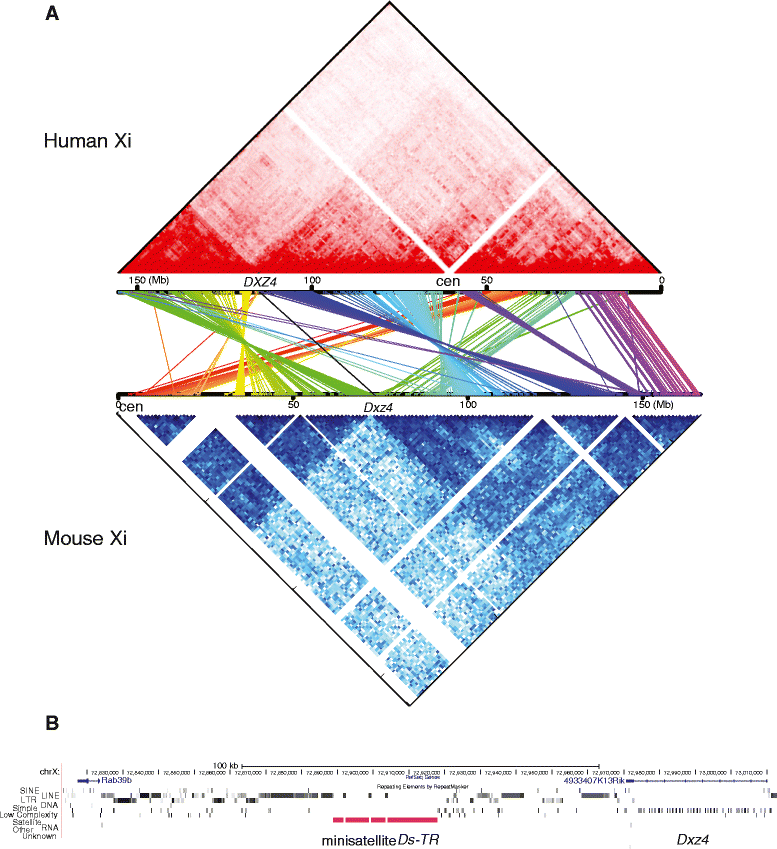
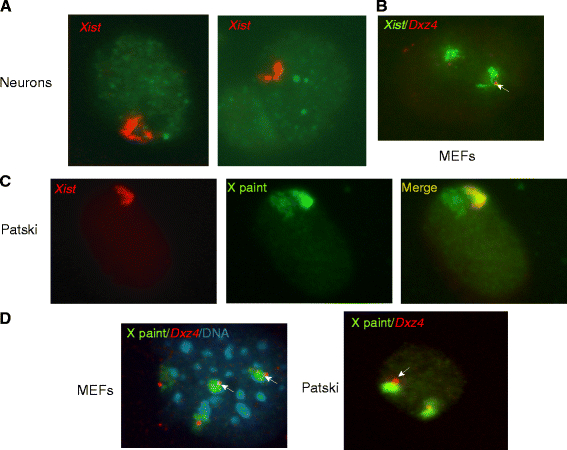
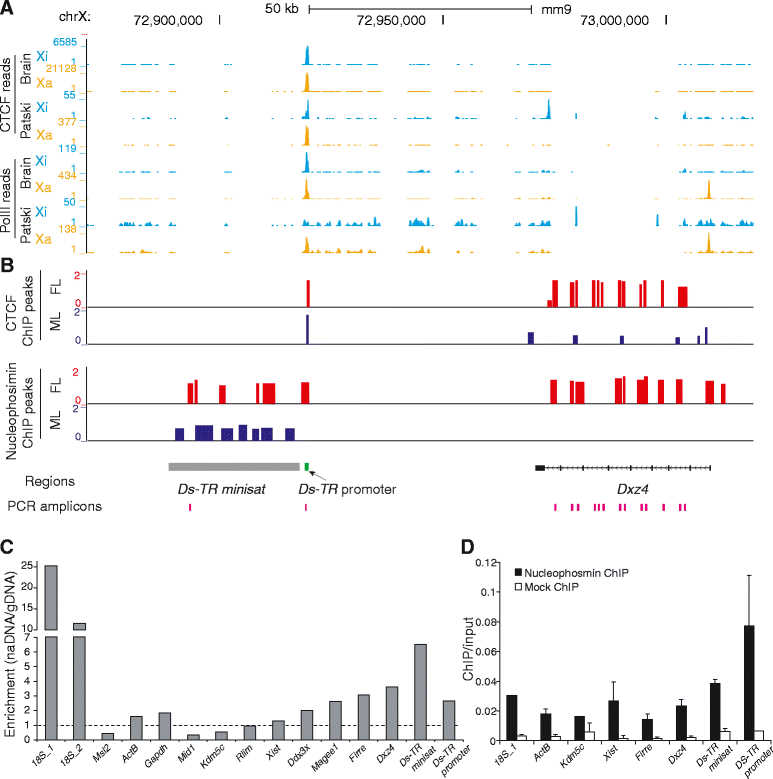
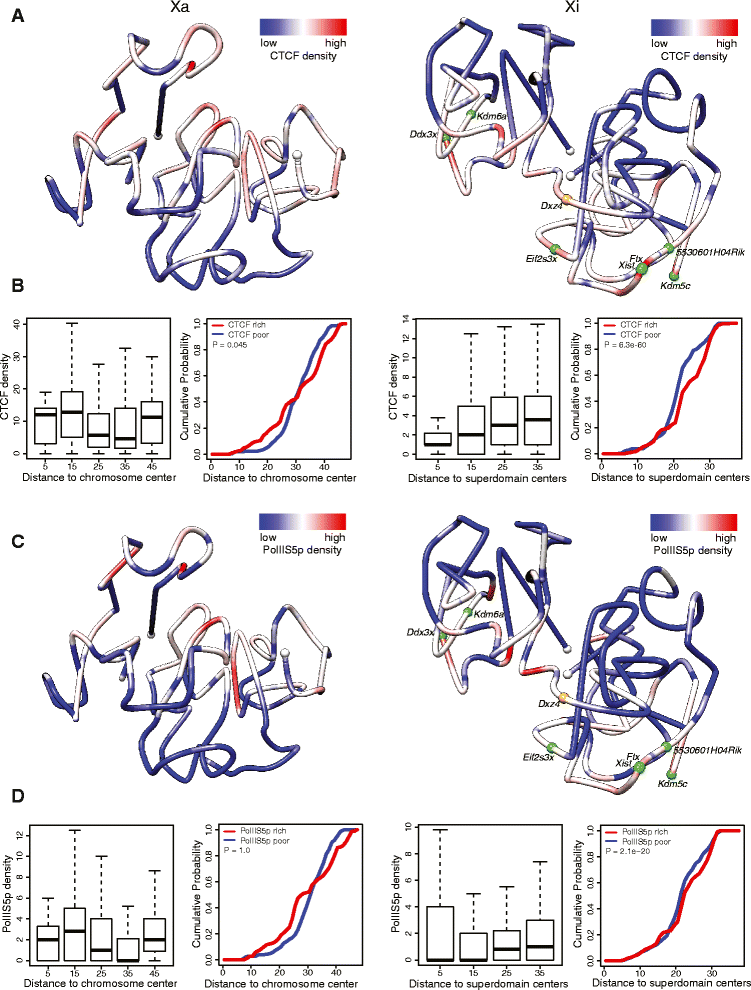
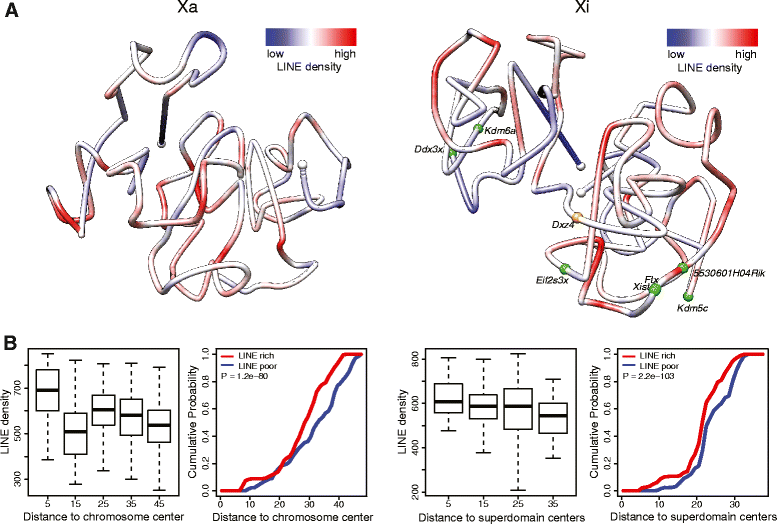
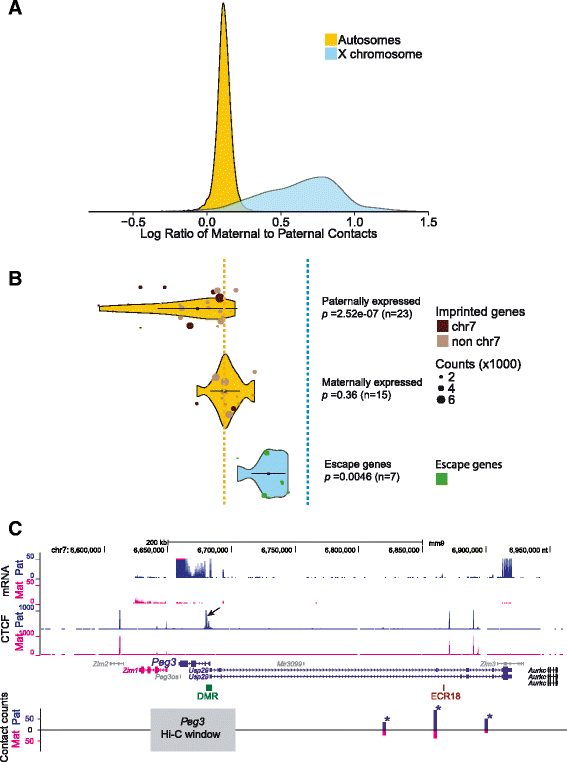
Results: We find radically different conformations for the two female mouse X chromosomes. The inactive X has two superdomains of frequent intrachromosomal contacts separated by a boundary region. Comparison with the recently reported two-superdomain structure of the human inactive X shows that the genomic content of the superdomains differs between species, but part of the boundary region is conserved and located near the Dxz4/DXZ4 locus. In mouse, the boundary region also contains a minisatellite, Ds-TR, and both Dxz4 and Ds-TR appear to be anchored to the nucleolus. Genes that escape X inactivation do not cluster but are located near the periphery of the 3D structure, as are regions enriched in CTCF or RNA polymerase. Fewer short-range intrachromosomal contacts are detected for the inactive alleles of genes subject to X inactivation compared with the active alleles and with genes that escape X inactivation. This pattern is also evident for imprinted genes, in which more chromatin contacts are detected for the expressed allele.
Conclusions: By applying a novel Hi-C method to map allelic chromatin contacts, we discover a specific bipartite organization of the mouse inactive X chromosome that probably plays an important role in maintenance of gene silencing.
Figures
Similar articles
-
Structural organization of the inactive X chromosome in the mouse.Nature. 2016 Jul 28;535(7613):575-9. doi: 10.1038/nature18589. Epub 2016 Jul 18. Nature. 2016. PMID: 27437574 Free PMC article.
-
Orientation-dependent Dxz4 contacts shape the 3D structure of the inactive X chromosome.Nat Commun. 2018 Apr 13;9(1):1445. doi: 10.1038/s41467-018-03694-y. Nat Commun. 2018. PMID: 29654302 Free PMC article.
-
Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture.Proc Natl Acad Sci U S A. 2016 Aug 2;113(31):E4504-12. doi: 10.1073/pnas.1609643113. Epub 2016 Jul 18. Proc Natl Acad Sci U S A. 2016. PMID: 27432957 Free PMC article.
-
Structural aspects of the inactive X chromosome.Philos Trans R Soc Lond B Biol Sci. 2017 Nov 5;372(1733):20160357. doi: 10.1098/rstb.2016.0357. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28947656 Free PMC article. Review.
-
X inactivation Xplained.Curr Opin Genet Dev. 2007 Oct;17(5):387-93. doi: 10.1016/j.gde.2007.08.001. Epub 2007 Sep 14. Curr Opin Genet Dev. 2007. PMID: 17869504 Review.
Cited by
-
Genomic identification and functional analysis of essential genes in Caenorhabditis elegans.BMC Genomics. 2018 Dec 4;19(1):871. doi: 10.1186/s12864-018-5251-3. BMC Genomics. 2018. PMID: 30514206 Free PMC article.
-
DeepLoop robustly maps chromatin interactions from sparse allele-resolved or single-cell Hi-C data at kilobase resolution.Nat Genet. 2022 Jul;54(7):1013-1025. doi: 10.1038/s41588-022-01116-w. Epub 2022 Jul 11. Nat Genet. 2022. PMID: 35817982 Free PMC article.
-
Female naïve human pluripotent stem cells carry X chromosomes with Xa-like and Xi-like folding conformations.Sci Adv. 2023 Aug 4;9(31):eadf2245. doi: 10.1126/sciadv.adf2245. Epub 2023 Aug 4. Sci Adv. 2023. PMID: 37540754 Free PMC article.
-
The role of epigenetics and chromatin structure in transcriptional regulation in malaria parasites.Brief Funct Genomics. 2019 Sep 24;18(5):302-313. doi: 10.1093/bfgp/elz005. Brief Funct Genomics. 2019. PMID: 31220857 Free PMC article. Review.
-
CTCF chromatin residence time controls three-dimensional genome organization, gene expression and DNA methylation in pluripotent cells.Nat Cell Biol. 2021 Aug;23(8):881-893. doi: 10.1038/s41556-021-00722-w. Epub 2021 Jul 29. Nat Cell Biol. 2021. PMID: 34326481
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

