The use of next generation sequencing in the diagnosis and typing of respiratory infections
- PMID: 26209388
- PMCID: PMC4533236
- DOI: 10.1016/j.jcv.2015.06.082
The use of next generation sequencing in the diagnosis and typing of respiratory infections
Erratum in
-
Erratum to "The use of next generation sequencing in the diagnosis and typing of respiratory infections"[J. Clin. Virol. 69 (2015) 96-100].J Clin Virol. 2015 Sep;70:128. doi: 10.1016/j.jcv.2015.06.101. Epub 2015 Jul 18. J Clin Virol. 2015. PMID: 28889994 Free PMC article. No abstract available.
Abstract
Background: Molecular assays are the gold standard methods used to diagnose viral respiratory pathogens. Pitfalls associated with this technique include limits to the number of targeted pathogens, the requirement for continuous monitoring to ensure sensitivity/specificity is maintained and the need to evolve to include emerging pathogens. Introducing target independent next generation sequencing (NGS) could resolve these issues and revolutionise respiratory viral diagnostics.
Objectives: To compare the sensitivity and specificity of target independent NGS against the current standard diagnostic test.
Study design: Diagnostic RT-PCR of clinical samples was carried out in parallel with target independent NGS. NGS sequences were analyzed to determine the proportion with viral origin and consensus sequences were used to establish viral genotypes and serotypes where applicable.
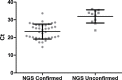
Results: 89 nasopharyngeal swabs were tested. A viral pathogen was detected in 43% of samples by NGS and 54% by RT-PCR. All NGS viral detections were confirmed by RT-PCR.
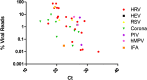
Conclusions: Target independent NGS can detect viral pathogens in clinical samples. Where viruses were detected by RT-PCR alone the Ct value was higher than those detected by both assays, suggesting an NGS detection cut-off - Ct=32. The sensitivity and specificity of NGS compared with RT-PCR was 78% and 80% respectively. This is lower than current diagnostic assays but NGS provided full genome sequences in some cases, allowing determination of viral subtype and serotype. Sequencing technology is improving rapidly and it is likely that within a short period of time sequencing depth will increase in-turn improving test sensitivity.
Keywords: Diagnostics; Next generation sequencing; Viral respiratory infection; Virus detection;.
Copyright © 2015 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Exploring the potential of next-generation sequencing in detection of respiratory viruses.J Clin Microbiol. 2014 Oct;52(10):3722-30. doi: 10.1128/JCM.01641-14. Epub 2014 Aug 6. J Clin Microbiol. 2014. PMID: 25100822 Free PMC article.
-
Direct multiplexed whole genome sequencing of respiratory tract samples reveals full viral genomic information.J Clin Virol. 2015 May;66:6-11. doi: 10.1016/j.jcv.2015.02.010. Epub 2015 Feb 18. J Clin Virol. 2015. PMID: 25866327 Free PMC article.
-
Application of next generation sequencing for the detection of human viral pathogens in clinical specimens.J Clin Virol. 2017 Jan;86:20-26. doi: 10.1016/j.jcv.2016.11.010. Epub 2016 Nov 22. J Clin Virol. 2017. PMID: 27902961 Free PMC article.
-
Next-generation sequencing technologies in diagnostic virology.J Clin Virol. 2013 Oct;58(2):346-50. doi: 10.1016/j.jcv.2013.03.003. Epub 2013 Mar 21. J Clin Virol. 2013. PMID: 23523339 Review.
-
Molecular diagnosis of respiratory virus infections.Crit Rev Clin Lab Sci. 2011 Sep-Dec;48(5-6):217-49. doi: 10.3109/10408363.2011.640976. Crit Rev Clin Lab Sci. 2011. PMID: 22185616 Review.
Cited by
-
Metagenomics Study of Viral Pathogens in Undiagnosed Respiratory Specimens and Identification of Human Enteroviruses at a Thailand Hospital.Am J Trop Med Hyg. 2016 Sep 7;95(3):663-669. doi: 10.4269/ajtmh.16-0062. Epub 2016 Jun 27. Am J Trop Med Hyg. 2016. PMID: 27352877 Free PMC article.
-
Advances in Clinical Sample Preparation for Identification and Characterization of Bacterial Pathogens Using Metagenomics.Front Public Health. 2018 Dec 12;6:363. doi: 10.3389/fpubh.2018.00363. eCollection 2018. Front Public Health. 2018. PMID: 30619804 Free PMC article. Review.
-
Selective Whole-Genome Amplification as a Tool to Enrich Specimens with Low Treponema pallidum Genomic DNA Copies for Whole-Genome Sequencing.mSphere. 2022 Jun 29;7(3):e0000922. doi: 10.1128/msphere.00009-22. Epub 2022 May 2. mSphere. 2022. PMID: 35491834 Free PMC article.
-
Rapid and precise diagnosis of disseminated T.marneffei infection assisted by high-throughput sequencing of multifarious specimens in a HIV-negative patient: a case report.BMC Infect Dis. 2018 Aug 7;18(1):379. doi: 10.1186/s12879-018-3276-5. BMC Infect Dis. 2018. PMID: 30086724 Free PMC article.
-
Composite Analysis of the Virome and Bacteriome of HIV/HPV Co-Infected Women Reveals Proxies for Immunodeficiency.Viruses. 2019 May 7;11(5):422. doi: 10.3390/v11050422. Viruses. 2019. PMID: 31067713 Free PMC article.
References
-
- Ip H.S. Identification and characterization of Highlands J virus from a Mississippi sandhill crane using unbiased next-generation sequencing. J. Virol. Methods. 2014;206:42–45. - PubMed
-
- Murdoch D.R. Effect of vitamin D3 supplementation on upper respiratory tract infections in healthy adults: the VIDARIS randomized controlled trial. JAMA. 2012;308(13):1333–1339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

