Two novel DXZ4-associated long noncoding RNAs show developmental changes in expression coincident with heterochromatin formation at the human (Homo sapiens) macrosatellite repeat
- PMID: 26188586
- PMCID: PMC4668219
- DOI: 10.1007/s10577-015-9479-3
Two novel DXZ4-associated long noncoding RNAs show developmental changes in expression coincident with heterochromatin formation at the human (Homo sapiens) macrosatellite repeat
Abstract
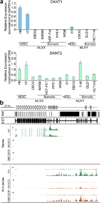
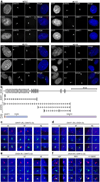
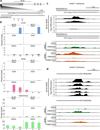
On the male X and female active X chromosome (Xa), the macrosatellite repeat (MSR) DXZ4 is packaged into constitutive heterochromatin characterized by CpG methylation and histone H3 tri-methylated at lysine-9 (H3K9me3). In contrast, DXZ4 on the female inactive X chromosome (Xi), is packaged into euchromatin, is bound by the architectural protein CCCTC-binding factor, and mediates Xi-specific long-range cis contact with similarly packaged tandem repeats on the Xi. In cancer, male DXZ4 can inappropriately revert to a Xi-like state and other MSRs have been reported to adopt alternate chromatin configurations in response to disease. Given this plasticity, we sought to identify factors that might control heterochromatin at DXZ4. In human embryonic stem cells, we found low levels of 5-hydroxymethylcytosine at DXZ4 and that this mark is lost upon differentiation as H3K9me3 is acquired. We identified two previously undescribed DXZ4 associated noncoding transcripts (DANT1 and DANT2) that are transcribed toward DXZ4 from promoters flanking the array. Each generates transcript isoforms that traverse the MSR. However, upon differentiation, enhancer of Zeste-2 silences DANT1, and DANT2 transcription terminates prior to entering DXZ4. These data support a model wherein DANT1 and/or DANT2 may function to regulate constitutive heterochromatin formation at this MSR.
Keywords: DXZ4; Euchromatin and heterochromatin; Human embryonic stem cells; Long noncoding RNA; Macrosatellite; X chromosome inactivation.
Conflict of interest statement
The author Debbie M. Figueroa declares they have no conflict of interest.
The author Emily M. Darrow declares they have no conflict of interest.
The author Brian P. Chadwick declares they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
Figures



Similar articles
-
DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts.Genome Res. 2008 Aug;18(8):1259-69. doi: 10.1101/gr.075713.107. Epub 2008 May 2. Genome Res. 2008. PMID: 18456864 Free PMC article.
-
The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome.Hum Mol Genet. 2012 Oct 15;21(20):4367-77. doi: 10.1093/hmg/dds270. Epub 2012 Jul 12. Hum Mol Genet. 2012. PMID: 22791747 Free PMC article.
-
The mouse DXZ4 homolog retains Ctcf binding and proximity to Pls3 despite substantial organizational differences compared to the primate macrosatellite.Genome Biol. 2012 Aug 20;13(8):R70. doi: 10.1186/gb-2012-13-8-r70. Genome Biol. 2012. PMID: 22906166 Free PMC article.
-
Modulation of Chromatin by Noncoding RNA.Annu Rev Genet. 2015;49:673-95. doi: 10.1146/annurev-genet-112414-055205. Annu Rev Genet. 2015. PMID: 26631517 Review.
-
The making of a Barr body: the mosaic of factors that eXIST on the mammalian inactive X chromosome.Biochem Cell Biol. 2016 Feb;94(1):56-70. doi: 10.1139/bcb-2015-0016. Epub 2015 Jun 24. Biochem Cell Biol. 2016. PMID: 26283003 Review.
Cited by
-
Pterostilbene leads to DNMT3B-mediated DNA methylation and silencing of OCT1-targeted oncogenes in breast cancer cells.J Nutr Biochem. 2021 Dec;98:108815. doi: 10.1016/j.jnutbio.2021.108815. Epub 2021 Jul 7. J Nutr Biochem. 2021. PMID: 34242723 Free PMC article.
-
Single-haplotype comparative genomics provides insights into lineage-specific structural variation during cat evolution.Nat Genet. 2023 Nov;55(11):1953-1963. doi: 10.1038/s41588-023-01548-y. Epub 2023 Nov 2. Nat Genet. 2023. PMID: 37919451 Free PMC article.
-
A novel long non-coding RNA from NBL2 pericentromeric macrosatellite forms a perinucleolar aggregate structure in colon cancer.Nucleic Acids Res. 2018 Jun 20;46(11):5504-5524. doi: 10.1093/nar/gky263. Nucleic Acids Res. 2018. PMID: 29912433 Free PMC article.
-
Forged by DXZ4, FIRRE, and ICCE: How Tandem Repeats Shape the Active and Inactive X Chromosome.Front Cell Dev Biol. 2020 Jan 21;7:328. doi: 10.3389/fcell.2019.00328. eCollection 2019. Front Cell Dev Biol. 2020. PMID: 32076600 Free PMC article.
-
Transcriptomic Analysis of Potential "lncRNA-mRNA" Interactions in Liver of the Marine Teleost Cynoglossus semilaevis Fed Diets With Different DHA/EPA Ratios.Front Physiol. 2019 Apr 2;10:331. doi: 10.3389/fphys.2019.00331. eCollection 2019. Front Physiol. 2019. PMID: 31001132 Free PMC article.
References
-
- Booth MJ, Branco MR, Ficz G, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336(6083):934–937. - PubMed
-
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13(1):7–13. - PubMed
-
- Brockdorff N, Ashworth A, Kay GF, et al. The product of the mouse Xist gene is a 15kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. - PubMed
-
- Brown CJ, Hendrich BD, Rupert JL, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

