Proteomics and Transcriptomics of BJAB Cells Expressing the Epstein-Barr Virus Noncoding RNAs EBER1 and EBER2
- PMID: 26121143
- PMCID: PMC4487896
- DOI: 10.1371/journal.pone.0124638
Proteomics and Transcriptomics of BJAB Cells Expressing the Epstein-Barr Virus Noncoding RNAs EBER1 and EBER2
Abstract
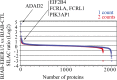
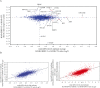
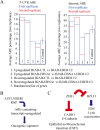
In Epstein-Barr virus (EBV) latent infection, the EBV-encoded RNAs EBER1 and EBER2 accumulate in the host cell nucleus to ~10(6) copies. While the expression of EBERs in cell lines is associated with transformation, a mechanistic explanation of their roles in EBV latency remains elusive. To identify EBER-specific gene expression features, we compared the proteome and mRNA transcriptome from BJAB cells (an EBV-negative B lymphoma cell line) stably transfected with an empty plasmid or with one carrying both EBER genes. We identified ~1800 proteins with at least 2 SILAC pair measurements, of which only 8 and 12 were up- and downregulated ≥ 2-fold, respectively. One upregulated protein was PIK3AP1, a B-cell specific protein adapter known to activate the PI3K-AKT signaling pathway, which regulates alternative splicing and translation in addition to its pro-survival effects. In the mRNA-seq data, the mRNA levels for some of the proteins changing in the SILAC data did not change. We instead observed isoform switch events. We validated the most relevant findings with biochemical assays. These corroborated the upregulation of PIK3AP1 and AKT activation in BJAB cells expressing high levels of both EBERs and EBNA1 (a surrogate of Burkitt's lymphoma EBV latency I) relative to those expressing only EBNA1. The mRNA-seq data in these cells showed multiple upregulated oncogenes whose mRNAs are enriched for 3´-UTR AU-rich elements (AREs), such as ccl3, ccr7, il10, vegfa and zeb1. The CCL3, CCR7, IL10 and VEGFA proteins promote cell proliferation and are associated with EBV-mediated lymphomas. In EBV latency, ZEB1 represses the transcription of ZEBRA, an EBV lytic phase activation factor. We previously found that EBER1 interacts with AUF1 in vivo and proposed stabilization of ARE-containing mRNAs. Thus, the ~10(6) copies of EBER1 may promote not only cell proliferation due to an increase in the levels of ARE-containing genes like ccl3, ccr7, il10, and vegfa, but also the maintenance of latency, through higher levels of zeb1.
Conflict of interest statement
Figures




Similar articles
-
Dominant-negative derivative of EBNA1 represses EBNA1-mediated transforming gene expression during the acute phase of Epstein-Barr virus infection independent of rapid loss of viral genome.Cancer Sci. 2010 Apr;101(4):876-81. doi: 10.1111/j.1349-7006.2009.01474.x. Epub 2009 Dec 16. Cancer Sci. 2010. PMID: 20132216 Free PMC article.
-
AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus.RNA. 2012 Nov;18(11):2073-82. doi: 10.1261/rna.034900.112. Epub 2012 Sep 25. RNA. 2012. PMID: 23012480 Free PMC article.
-
A Polymorphism in the Epstein-Barr Virus EBER2 Noncoding RNA Drives In Vivo Expansion of Latently Infected B Cells.mBio. 2022 Jun 28;13(3):e0083622. doi: 10.1128/mbio.00836-22. Epub 2022 Jun 1. mBio. 2022. PMID: 35642944 Free PMC article.
-
Epstein-Barr virus-encoded EBNA1 and ZEBRA: targets for therapeutic strategies against EBV-carrying cancers.J Pathol. 2015 Jan;235(2):334-41. doi: 10.1002/path.4431. J Pathol. 2015. PMID: 25186125 Review.
-
Multifunctional non-coding Epstein-Barr virus encoded RNAs (EBERs) contribute to viral pathogenesis.Virus Res. 2016 Jan 2;212:30-8. doi: 10.1016/j.virusres.2015.08.007. Epub 2015 Aug 18. Virus Res. 2016. PMID: 26292159 Review.
Cited by
-
Consideration of Epstein-Barr Virus-Encoded Noncoding RNAs EBER1 and EBER2 as a Functional Backup of Viral Oncoprotein Latent Membrane Protein 1.mBio. 2016 Jan 19;7(1):e01926-15. doi: 10.1128/mBio.01926-15. mBio. 2016. PMID: 26787829 Free PMC article.
-
Transcriptome-wide analysis of alternative RNA splicing events in Epstein-Barr virus-associated gastric carcinomas.PLoS One. 2017 May 11;12(5):e0176880. doi: 10.1371/journal.pone.0176880. eCollection 2017. PLoS One. 2017. PMID: 28493890 Free PMC article.
-
Global Profiling of the Cellular Alternative RNA Splicing Landscape during Virus-Host Interactions.PLoS One. 2016 Sep 6;11(9):e0161914. doi: 10.1371/journal.pone.0161914. eCollection 2016. PLoS One. 2016. PMID: 27598998 Free PMC article.
-
The Epstein-Barr virus noncoding RNA EBER2 transactivates the UCHL1 deubiquitinase to accelerate cell growth.Proc Natl Acad Sci U S A. 2021 Oct 26;118(43):e2115508118. doi: 10.1073/pnas.2115508118. Proc Natl Acad Sci U S A. 2021. PMID: 34686609 Free PMC article.
-
Viral modulation of cellular RNA alternative splicing: A new key player in virus-host interactions?Wiley Interdiscip Rev RNA. 2019 Sep;10(5):e1543. doi: 10.1002/wrna.1543. Epub 2019 Apr 29. Wiley Interdiscip Rev RNA. 2019. PMID: 31034770 Free PMC article. Review.
References
-
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4(10):757–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

