Locating Herpesvirus Bcl-2 Homologs in the Specificity Landscape of Anti-Apoptotic Bcl-2 Proteins
- PMID: 26009469
- PMCID: PMC4520770
- DOI: 10.1016/j.jmb.2015.05.015
Locating Herpesvirus Bcl-2 Homologs in the Specificity Landscape of Anti-Apoptotic Bcl-2 Proteins
Abstract
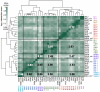
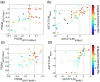
Viral homologs of the anti-apoptotic Bcl-2 proteins are highly diverged from their mammalian counterparts, yet they perform overlapping functions by binding and inhibiting BH3 (Bcl-2 homology 3)-motif-containing proteins. We investigated the BH3 binding properties of the herpesvirus Bcl-2 homologs KSBcl-2, BHRF1, and M11, as they relate to those of the human Bcl-2 homologs Mcl-1, Bfl-1, Bcl-w, Bcl-xL, and Bcl-2. Analysis of the sequence and structure of the BH3 binding grooves showed that, despite low sequence identity, M11 has structural similarities to Bcl-xL, Bcl-2, and Bcl-w. BHRF1 and KSBcl-2 are more structurally similar to Mcl-1 than to the other human proteins. Binding to human BH3-like peptides showed that KSBcl-2 has similar specificity to Mcl-1, and BHRF1 has a restricted binding profile; M11 binding preferences are distinct from those of Bcl-xL, Bcl-2, and Bcl-w. Because KSBcl-2 and BHRF1 are from human herpesviruses associated with malignancies, we screened computationally designed BH3 peptide libraries using bacterial surface display to identify selective binders of KSBcl-2 or BHRF1. The resulting peptides bound to KSBcl-2 and BHRF1 in preference to Bfl-1, Bcl-w, Bcl-xL, and Bcl-2 but showed only modest specificity over Mcl-1. Rational mutagenesis increased specificity against Mcl-1, resulting in a peptide with a dissociation constant of 2.9nM for binding to KSBcl-2 and >1000-fold specificity over other Bcl-2 proteins, as well as a peptide with >70-fold specificity for BHRF1. In addition to providing new insights into viral Bcl-2 binding specificity, this study will inform future work analyzing the interaction properties of homologous binding domains and designing specific protein interaction partners.
Keywords: BH3 peptides; BHRF1; KSBcl-2; bacterial surface display; peptide design.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures







Similar articles
-
A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak.Proc Natl Acad Sci U S A. 1997 Jan 21;94(2):690-4. doi: 10.1073/pnas.94.2.690. Proc Natl Acad Sci U S A. 1997. PMID: 9012846 Free PMC article.
-
Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL.J Mol Biol. 2010 May 21;398(5):747-62. doi: 10.1016/j.jmb.2010.03.058. Epub 2010 Apr 2. J Mol Biol. 2010. PMID: 20363230 Free PMC article.
-
Structural basis for the conserved binding mechanism of MDM2-inhibiting peptides and anti-apoptotic Bcl-2 family proteins.Biochem Biophys Res Commun. 2014 Feb 28;445(1):120-5. doi: 10.1016/j.bbrc.2014.01.130. Epub 2014 Feb 1. Biochem Biophys Res Commun. 2014. PMID: 24491548
-
Structural biology of the Bcl-2 family of proteins.Biochim Biophys Acta. 2004 Mar 1;1644(2-3):83-94. doi: 10.1016/j.bbamcr.2003.08.012. Biochim Biophys Acta. 2004. PMID: 14996493 Review.
-
Functional similarity between adenovirus E1B 19-kDa protein and proteins encoded by Bcl-2 proto-oncogene and Epstein-Barr virus BHRF1 gene.Curr Top Microbiol Immunol. 1995;199 ( Pt 1):153-61. doi: 10.1007/978-3-642-79496-4_9. Curr Top Microbiol Immunol. 1995. PMID: 7555053 Review. No abstract available.
Cited by
-
Rapid Evaluation of Staple Placement in Stabilized α Helices Using Bacterial Surface Display.ACS Chem Biol. 2023 Apr 21;18(4):905-914. doi: 10.1021/acschembio.3c00048. Epub 2023 Apr 11. ACS Chem Biol. 2023. PMID: 37039514 Free PMC article.
-
Epistatic mutations in PUMA BH3 drive an alternate binding mode to potently and selectively inhibit anti-apoptotic Bfl-1.Elife. 2017 Jun 8;6:e25541. doi: 10.7554/eLife.25541. Elife. 2017. PMID: 28594323 Free PMC article.
-
Transient Unfolding and Long-Range Interactions in Viral BCL2 M11 Enable Binding to the BECN1 BH3 Domain.Biomolecules. 2020 Sep 11;10(9):1308. doi: 10.3390/biom10091308. Biomolecules. 2020. PMID: 32932757 Free PMC article.
-
Enriching Peptide Libraries for Binding Affinity and Specificity Through Computationally Directed Library Design.Methods Mol Biol. 2017;1561:213-232. doi: 10.1007/978-1-4939-6798-8_13. Methods Mol Biol. 2017. PMID: 28236241 Free PMC article.
-
[Effects of simvastatin on aortic vascular endothelial cell apoptosis and Bcl-2 protein expression in a rat model of atherosclerosis].Nan Fang Yi Ke Da Xue Xue Bao. 2017 Nov 20;37(11):1456-1460. doi: 10.3969/j.issn.1673-4254.2017.11.05. Nan Fang Yi Ke Da Xue Xue Bao. 2017. PMID: 29180324 Free PMC article. Chinese.
References
-
- Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. - PubMed
-
- Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

