The 3D structure of Kaposi sarcoma herpesvirus LANA C-terminal domain bound to DNA
- PMID: 25947153
- PMCID: PMC4450395
- DOI: 10.1073/pnas.1421804112
The 3D structure of Kaposi sarcoma herpesvirus LANA C-terminal domain bound to DNA
Abstract
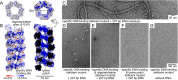
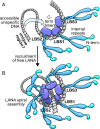
Kaposi sarcoma herpesvirus (KSHV) persists as a latent nuclear episome in dividing host cells. This episome is tethered to host chromatin to ensure proper segregation during mitosis. For duplication of the latent genome, the cellular replication machinery is recruited. Both of these functions rely on the constitutively expressed latency-associated nuclear antigen (LANA) of the virus. Here, we report the crystal structure of the KSHV LANA DNA-binding domain (DBD) in complex with its high-affinity viral target DNA, LANA binding site 1 (LBS1), at 2.9 Å resolution. In contrast to homologous proteins such as Epstein-Barr virus nuclear antigen 1 (EBNA-1) of the related γ-herpesvirus Epstein-Barr virus, specific DNA recognition by LANA is highly asymmetric. In addition to solving the crystal structure, we found that apart from the two known LANA binding sites, LBS1 and LBS2, LANA also binds to a novel site, denoted LBS3. All three sites are located in a region of the KSHV terminal repeat subunit previously recognized as a minimal replicator. Moreover, we show that the LANA DBD can coat DNA of arbitrary sequence by virtue of a characteristic lysine patch, which is absent in EBNA-1 of the Epstein-Barr virus. Likely, these higher-order assemblies involve the self-association of LANA into supermolecular spirals. One such spiral assembly was solved as a crystal structure of 3.7 Å resolution in the absence of DNA. On the basis of our data, we propose a model for the controlled nucleation of higher-order LANA oligomers that might contribute to the characteristic subnuclear KSHV microdomains ("LANA speckles"), a hallmark of KSHV latency.
Keywords: DNA-binding protein; KSHV LANA; X-ray crystallography; gammaherpesvirinae; viral latency.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Site-specific association with host and viral chromatin by Kaposi's sarcoma-associated herpesvirus LANA and its reversal during lytic reactivation.J Virol. 2014 Jun;88(12):6762-77. doi: 10.1128/JVI.00268-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696474 Free PMC article.
-
The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells.J Virol. 2002 Nov;76(22):11677-87. doi: 10.1128/jvi.76.22.11677-11687.2002. J Virol. 2002. PMID: 12388727 Free PMC article.
-
Mutational analysis of the latency-associated nuclear antigen DNA-binding domain of Kaposi's sarcoma-associated herpesvirus reveals structural conservation among gammaherpesvirus origin-binding proteins.J Gen Virol. 2010 Sep;91(Pt 9):2203-15. doi: 10.1099/vir.0.020958-0. Epub 2010 May 19. J Gen Virol. 2010. PMID: 20484563 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
Cited by
-
Epstein-Barr virus enhances genome maintenance of Kaposi sarcoma-associated herpesvirus.Proc Natl Acad Sci U S A. 2018 Nov 27;115(48):E11379-E11387. doi: 10.1073/pnas.1810128115. Epub 2018 Nov 14. Proc Natl Acad Sci U S A. 2018. PMID: 30429324 Free PMC article.
-
Visualization of molecular biology: The LANA tether.Proc Natl Acad Sci U S A. 2018 May 8;115(19):4816-4818. doi: 10.1073/pnas.1804797115. Epub 2018 Apr 24. Proc Natl Acad Sci U S A. 2018. PMID: 29691321 Free PMC article. No abstract available.
-
The Kaposi Sarcoma Herpesvirus Latency-associated Nuclear Antigen DNA Binding Domain Dorsal Positive Electrostatic Patch Facilitates DNA Replication and Episome Persistence.J Biol Chem. 2015 Nov 20;290(47):28084-28096. doi: 10.1074/jbc.M115.674622. Epub 2015 Sep 29. J Biol Chem. 2015. PMID: 26420481 Free PMC article.
-
Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS.Proc Natl Acad Sci U S A. 2016 Feb 23;113(8):E1034-43. doi: 10.1073/pnas.1516812113. Epub 2016 Jan 25. Proc Natl Acad Sci U S A. 2016. PMID: 26811480 Free PMC article.
-
Recent advances in understanding Kaposi's sarcoma-associated herpesvirus.F1000Res. 2016 Apr 25;5:F1000 Faculty Rev-740. doi: 10.12688/f1000research.7612.1. eCollection 2016. F1000Res. 2016. PMID: 27158465 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

