The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer
- PMID: 25653311
- PMCID: PMC4381521
- DOI: 10.1101/gr.185926.114
The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer
Abstract
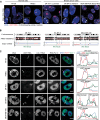
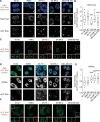
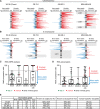
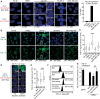
Disappearance of the Barr body is considered a hallmark of cancer, although whether this corresponds to genetic loss or to epigenetic instability and transcriptional reactivation is unclear. Here we show that breast tumors and cell lines frequently display major epigenetic instability of the inactive X chromosome, with highly abnormal 3D nuclear organization and global perturbations of heterochromatin, including gain of euchromatic marks and aberrant distributions of repressive marks such as H3K27me3 and promoter DNA methylation. Genome-wide profiling of chromatin and transcription reveal modified epigenomic landscapes in cancer cells and a significant degree of aberrant gene activity from the inactive X chromosome, including several genes involved in cancer promotion. We demonstrate that many of these genes are aberrantly reactivated in primary breast tumors, and we further demonstrate that epigenetic instability of the inactive X can lead to perturbed dosage of X-linked factors. Taken together, our study provides the first integrated analysis of the inactive X chromosome in the context of breast cancer and establishes that epigenetic erosion of the inactive X can lead to the disappearance of the Barr body in breast cancer cells. This work offers new insights and opens up the possibility of exploiting the inactive X chromosome as an epigenetic biomarker at the molecular and cytological levels in cancer.
© 2015 Chaligné et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Similar articles
-
The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation.Genome Biol. 2015 Mar 12;16(1):52. doi: 10.1186/s13059-015-0618-0. Genome Biol. 2015. PMID: 25887447 Free PMC article.
-
Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome.Hum Mol Genet. 2003 Sep 1;12(17):2167-78. doi: 10.1093/hmg/ddg229. Epub 2003 Jul 15. Hum Mol Genet. 2003. PMID: 12915472
-
Uncoupling of X-linked gene silencing from XIST binding by DICER1 and chromatin modulation on human inactive X chromosome.Chromosoma. 2015 Jun;124(2):249-62. doi: 10.1007/s00412-014-0495-4. Epub 2014 Nov 27. Chromosoma. 2015. PMID: 25428210
-
RNA and protein actors in X-chromosome inactivation.Cold Spring Harb Symp Quant Biol. 2006;71:419-28. doi: 10.1101/sqb.2006.71.058. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381324 Review.
-
The making of a Barr body: the mosaic of factors that eXIST on the mammalian inactive X chromosome.Biochem Cell Biol. 2016 Feb;94(1):56-70. doi: 10.1139/bcb-2015-0016. Epub 2015 Jun 24. Biochem Cell Biol. 2016. PMID: 26283003 Review.
Cited by
-
trans-Acting Factors and cis Elements Involved in the Human Inactive X Chromosome Organization and Compaction.Genet Res (Camb). 2021 May 7;2021:6683460. doi: 10.1155/2021/6683460. eCollection 2021. Genet Res (Camb). 2021. PMID: 34035662 Free PMC article. Review.
-
A new sex-specific underlying mechanism for female schizophrenia: accelerated skewed X chromosome inactivation.Biol Sex Differ. 2020 Jul 17;11(1):39. doi: 10.1186/s13293-020-00315-6. Biol Sex Differ. 2020. PMID: 32680558 Free PMC article.
-
Maintenance of epigenetic landscape requires CIZ1 and is corrupted in differentiated fibroblasts in long-term culture.Nat Commun. 2019 Jan 28;10(1):460. doi: 10.1038/s41467-018-08072-2. Nat Commun. 2019. PMID: 30692537 Free PMC article.
-
Partially methylated domains are hypervariable in breast cancer and fuel widespread CpG island hypermethylation.Nat Commun. 2019 Apr 15;10(1):1749. doi: 10.1038/s41467-019-09828-0. Nat Commun. 2019. PMID: 30988298 Free PMC article.
-
Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis.Cancer Res. 2018 Aug 1;78(15):4316-4330. doi: 10.1158/0008-5472.CAN-18-1102. Epub 2018 Jul 19. Cancer Res. 2018. PMID: 30026327 Free PMC article.
References
-
- Azoulay S, Laé M, Fréneaux P, Merle S, Al Ghuzlan A, Chnecker C, Rosty C, Klijanienko J, Sigal-Zafrani B, Salmon R, et al.2005. KIT is highly expressed in adenoid cystic carcinoma of the breast, a basal-like carcinoma associated with a favorable outcome. Mod Pathol 18: 1623–1631. - PubMed
-
- Barr ML, Moore KL. 1957. Chromosomes, sex chromatin, and cancer. Proc Can Cancer Conf 2: 3–16. - PubMed
-
- Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet 27: 20–21. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al.2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
