De novo isolation of antibodies with pH-dependent binding properties
- PMID: 25608219
- PMCID: PMC4623423
- DOI: 10.1080/19420862.2015.1006993
De novo isolation of antibodies with pH-dependent binding properties
Abstract
pH-dependent antibodies are engineered to release their target at a slightly acidic pH, a property making them suitable for clinical as well as biotechnological applications. Such antibodies were previously obtained by histidine scanning of pre-existing antibodies, a labor-intensive strategy resulting in antibodies that displayed residual binding to their target at pH 6.0. We report here the de novo isolation of pH-dependent antibodies selected by phage display from libraries enriched in histidines. Strongly pH-dependent clones with various affinity profiles against CXCL10 were isolated by this method. Our best candidate has nanomolar affinity for CXCL10 at pH 7.2, but no residual binding was detected at pH 6.0. We therefore propose that this new process is an efficient strategy to generate pH-dependent antibodies.
Keywords: BLI, bio-layer interferometry; CDR, complementary determining region; CDRH, CDR of the heavy chain; CDRL, CDR of the light chain; ELISA, enzyme-linked immunosorbent assay; GPCR, G protein-coupled receptor; KB, kinetic buffer; PBS, phosphate buffered saline; SPR, surface plasmon resonance; antibody recycling; chemokine; histidine; mAb, monoclonal antibody; monoclonal antibody; pH-dependency; phage display; phage libraries; scFv, single-chain variable fragment.
Figures
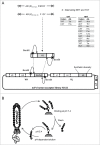
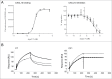


Similar articles
-
A combination of in vitro techniques for efficient discovery of functional monoclonal antibodies against human CXC chemokine receptor-2 (CXCR2).MAbs. 2014;6(6):1415-24. doi: 10.4161/mabs.36237. MAbs. 2014. PMID: 25484047 Free PMC article.
-
Construction and molecular characterization of mouse single-chain variable fragment antibodies against Burkholderia mallei and Burkholderia pseudomallei.J Immunol Methods. 2011 Feb 28;365(1-2):101-9. doi: 10.1016/j.jim.2010.12.004. Epub 2010 Dec 21. J Immunol Methods. 2011. PMID: 21172353
-
Combining phage display with de novo protein sequencing for reverse engineering of monoclonal antibodies.MAbs. 2016;8(3):501-12. doi: 10.1080/19420862.2016.1145865. MAbs. 2016. PMID: 26852694 Free PMC article.
-
Generation and analysis of the improved human HAL9/10 antibody phage display libraries.BMC Biotechnol. 2015 Feb 19;15:10. doi: 10.1186/s12896-015-0125-0. BMC Biotechnol. 2015. PMID: 25888378 Free PMC article.
-
A generic approach to engineer antibody pH-switches using combinatorial histidine scanning libraries and yeast display.MAbs. 2015;7(1):138-51. doi: 10.4161/19420862.2014.985993. MAbs. 2015. PMID: 25523975 Free PMC article.
Cited by
-
Evolution of phage display libraries for therapeutic antibody discovery.MAbs. 2023 Jan-Dec;15(1):2213793. doi: 10.1080/19420862.2023.2213793. MAbs. 2023. PMID: 37222232 Free PMC article. Review.
-
Structure-based engineering of pH-dependent antibody binding for selective targeting of solid-tumor microenvironment.MAbs. 2020 Jan-Dec;12(1):1682866. doi: 10.1080/19420862.2019.1682866. MAbs. 2020. PMID: 31777319 Free PMC article.
-
Elimination of plasma soluble antigen in cynomolgus monkeys by combining pH-dependent antigen binding and novel Fc engineering.MAbs. 2022 Jan-Dec;14(1):2068213. doi: 10.1080/19420862.2022.2068213. MAbs. 2022. PMID: 35482905 Free PMC article.
-
A stepwise mutagenesis approach using histidine and acidic amino acid to engineer highly pH-dependent protein switches.3 Biotech. 2022 Jan;12(1):21. doi: 10.1007/s13205-021-03079-x. Epub 2021 Dec 20. 3 Biotech. 2022. PMID: 34956814 Free PMC article.
-
Engineered IgG1-Fc--one fragment to bind them all.Immunol Rev. 2016 Mar;270(1):113-31. doi: 10.1111/imr.12385. Immunol Rev. 2016. PMID: 26864108 Free PMC article. Review.
References
-
- Yagami H, Kato H, Tsumoto K, Tomita M. Monoclonal antibodies based on hybridoma technology. Pharm Pat Anal 2013; 2:249-63; http://dx.doi.org/10.4155/ppa.13.2 - DOI - PubMed
-
- Hairul Bahara NH, Tye GJ, Choong YS, Ong EB, Ismail A, Lim TS. Phage display antibodies for diagnostic applications. Biologicals 2013; 41:209-16; PMID:23647952; http://dx.doi.org/10.1016/j.biologicals.2013.04.001 - DOI - PubMed
-
- Vincent KJ, Zurini M. Current strategies in antibody engineering: Fc engineering and pH-dependent antigen binding, bispecific antibodies and antibody drug conjugates. Biotechnol J 2012; 7:1444-50; PMID:23125076; http://dx.doi.org/10.1002/biot.201200250 - DOI - PubMed
-
- Sela-Culang I, Kunik V, Ofran Y. The structural basis of antibody-antigen recognition. Front Immunol 2013; 4:302; PMID:24115948; http://dx.doi.org/10.3389/fimmu.2013.00302 - DOI - PMC - PubMed
-
- Aggarwal RS. What's fueling the biotech engine-2012 to 2013. Nat Biotechnol 2014; 32:32-39; PMID:24406926; http://dx.doi.org/10.1038/nbt.2794 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
