Comprehensive review of human sapoviruses
- PMID: 25567221
- PMCID: PMC4284302
- DOI: 10.1128/CMR.00011-14
Comprehensive review of human sapoviruses
Abstract
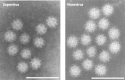
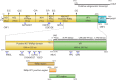
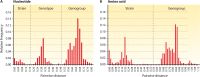
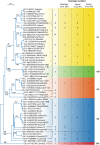
Sapoviruses cause acute gastroenteritis in humans and animals. They belong to the genus Sapovirus within the family Caliciviridae. They infect and cause disease in humans of all ages, in both sporadic cases and outbreaks. The clinical symptoms of sapovirus gastroenteritis are indistinguishable from those caused by noroviruses, so laboratory diagnosis is essential to identify the pathogen. Sapoviruses are highly diverse genetically and antigenically. Currently, reverse transcription-PCR (RT-PCR) assays are widely used for sapovirus detection from clinical specimens due to their high sensitivity and broad reactivity as well as the lack of sensitive assays for antigen detection or cell culture systems for the detection of infectious viruses. Sapoviruses were first discovered in 1976 by electron microscopy in diarrheic samples of humans. To date, sapoviruses have also been detected from several animals: pigs, mink, dogs, sea lions, and bats. In this review, we focus on genomic and antigenic features, molecular typing/classification, detection methods, and clinical and epidemiological profiles of human sapoviruses.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
[Genetic diversity of human caliciviruses detected in schoolchildren with gastroenteritis in Nizhni Novgorod].Vopr Virusol. 2009 Nov-Dec;54(6):24-8. Vopr Virusol. 2009. PMID: 20030278 Russian.
-
Polymerase chain reaction primer sets for the detection of genetically diverse human sapoviruses.Arch Virol. 2020 Oct;165(10):2335-2340. doi: 10.1007/s00705-020-04746-9. Epub 2020 Jul 27. Arch Virol. 2020. PMID: 32719956 Free PMC article.
-
Distribution and Genetic Variability of Sapoviruses in Africa.Viruses. 2020 Apr 27;12(5):490. doi: 10.3390/v12050490. Viruses. 2020. PMID: 32349380 Free PMC article. Review.
-
Prevalence and molecular characterization of human noroviruses and sapoviruses in Ethiopia.Arch Virol. 2016 Aug;161(8):2169-82. doi: 10.1007/s00705-016-2887-7. Epub 2016 May 19. Arch Virol. 2016. PMID: 27193022
-
Porcine sapoviruses: Pathogenesis, epidemiology, genetic diversity, and diagnosis.Virus Res. 2020 Sep;286:198025. doi: 10.1016/j.virusres.2020.198025. Epub 2020 May 26. Virus Res. 2020. PMID: 32470356 Free PMC article. Review.
Cited by
-
Propagating and banking genetically diverse human sapovirus strains using a human duodenal cell line: investigating antigenic differences between strains.J Virol. 2024 Sep 17;98(9):e0063924. doi: 10.1128/jvi.00639-24. Epub 2024 Aug 12. J Virol. 2024. PMID: 39132992
-
Comprehensive Genomics Investigation of Neboviruses Reveals Distinct Codon Usage Patterns and Host Specificity.Microorganisms. 2024 Mar 29;12(4):696. doi: 10.3390/microorganisms12040696. Microorganisms. 2024. PMID: 38674640 Free PMC article.
-
Clinical severity of enteric viruses detected using a quantitative molecular assay compared to conventional assays in the Global Enteric Multicenter Study.J Infect Dis. 2024 Apr 18:jiae201. doi: 10.1093/infdis/jiae201. Online ahead of print. J Infect Dis. 2024. PMID: 38637321
-
Detection and complete genome characterization of a genogroup X (GX) sapovirus (family Caliciviridae) from a golden jackal (Canis aureus) in Hungary.Arch Virol. 2024 Apr 17;169(5):100. doi: 10.1007/s00705-024-06034-2. Arch Virol. 2024. PMID: 38630394 Free PMC article.
-
Sapovirus: an emerging pathogen in kidney transplant recipients?Infection. 2024 Oct;52(5):1831-1838. doi: 10.1007/s15010-024-02242-9. Epub 2024 Apr 9. Infection. 2024. PMID: 38592660 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

