Kaposi's sarcoma-associated herpesvirus genome programming during the early stages of primary infection of peripheral blood mononuclear cells
- PMID: 25516617
- PMCID: PMC4271552
- DOI: 10.1128/mBio.02261-14
Kaposi's sarcoma-associated herpesvirus genome programming during the early stages of primary infection of peripheral blood mononuclear cells
Abstract
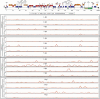
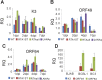
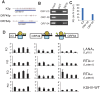
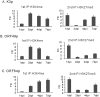
The early period of Kaposi's sarcoma-associated herpesvirus (KSHV) infection involves the dynamic expression of viral genes, which are temporally and epigenetically regulated. KSHV can effectively infect and persist in endothelial as well as human B cells with different gene expression patterns. To understand the temporal epigenetic changes which occur when KSHV infects the lymphocytic compartment, we infected human peripheral blood mononuclear cells (PBMCs) and comprehensively analyzed the changes which occurred at the binding sites of virally encoded lytic as well as latent proteins along with epigenetic modifications across the KSHV genome during early primary infection. Using chromatin immunoprecipitation (ChIP) assays, we showed that the KSHV genome acquires a uniquely distinct histone modification pattern of methylation (H3K4me3, H3K9me3, and H3K27me3) and acetylation (H3Ac) during de novo infection of human PBMCs. This pattern showed that the epigenetic changes were temporally controlled. The binding profiles of KSHV latent protein LANA and the immediate early proteins RTA and K8 showed specific patterns at different times postinfection, which reflects the gene expression program. Further analysis demonstrated that KSHV can concurrently express lytic and latent genes which were associated with histone modifications at these specific regions on the viral genome. We identified three KSHV genes, K3, ORF49, and ORF64, which exhibited different profiles of histone modifications during the early stages of PBMC infection. These studies established a distinct pattern of epigenetic modification which correlates with viral gene expression temporally regulated during the first 7 days of PBMC infection and provides clues to the regulatory program required for successful infection by KSHV of human PBMCs.
Importance: Kaposi's sarcoma-associated herpesvirus (KSHV) has been documented as one of the major contributors to morbidity and mortality in AIDS patients during the AIDS pandemic. During its life cycle, KSHV undergoes latent and lytic replication. Typically, KSHV maintains a stringent preference for latent infection in the infected B cells. However, 1 to 5% of infected cells undergo spontaneous lytic reactivation. KSHV lytic replication and infection of new cells are likely to be critical for maintaining the population of infected cells which drive virus-associated pathogenesis. Here, we explored the temporal changes of crucial histone marks on the KSHV genome during early infection of human primary peripheral blood mononuclear cells (PBMCs), which are a physiologically relevant system for monitoring primary infection. These results showed that KSHV possessed a distinct pattern of epigenetic marks during early infection of PBMCs. Further, KSHV concurrently expressed lytic and latent genes during this early period. These results now provide new evidence which contributes to understanding the molecular mechanism that regulates viral gene expression during early infection.
Copyright © 2014 Jha et al.
Figures


Similar articles
-
Transcriptome analysis of Kaposi's sarcoma-associated herpesvirus during de novo primary infection of human B and endothelial cells.J Virol. 2015 Mar;89(6):3093-111. doi: 10.1128/JVI.02507-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552714 Free PMC article.
-
Activation of Kaposi's sarcoma-associated herpesvirus (KSHV) by inhibitors of class III histone deacetylases: identification of sirtuin 1 as a regulator of the KSHV life cycle.J Virol. 2014 Jun;88(11):6355-67. doi: 10.1128/JVI.00219-14. Epub 2014 Mar 26. J Virol. 2014. PMID: 24672028 Free PMC article.
-
Epigenetic analysis of KSHV latent and lytic genomes.PLoS Pathog. 2010 Jul 22;6(7):e1001013. doi: 10.1371/journal.ppat.1001013. PLoS Pathog. 2010. PMID: 20661424 Free PMC article.
-
Molecular biology of KSHV lytic reactivation.Viruses. 2015 Jan 14;7(1):116-53. doi: 10.3390/v7010116. Viruses. 2015. PMID: 25594835 Free PMC article. Review.
-
Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus replication.Semin Cancer Biol. 2009 Jun;19(3):153-7. doi: 10.1016/j.semcancer.2009.02.010. Epub 2009 Feb 21. Semin Cancer Biol. 2009. PMID: 19429478 Free PMC article. Review.
Cited by
-
MLL1 is regulated by KSHV LANA and is important for virus latency.Nucleic Acids Res. 2021 Dec 16;49(22):12895-12911. doi: 10.1093/nar/gkab1094. Nucleic Acids Res. 2021. PMID: 34850113 Free PMC article.
-
Gammaherpesvirus Infection of Human Neuronal Cells.mBio. 2015 Dec 1;6(6):e01844-15. doi: 10.1128/mBio.01844-15. mBio. 2015. PMID: 26628726 Free PMC article.
-
Kaposi sarcoma-associated herpesvirus: immunobiology, oncogenesis, and therapy.J Clin Invest. 2016 Sep 1;126(9):3165-75. doi: 10.1172/JCI84418. Epub 2016 Sep 1. J Clin Invest. 2016. PMID: 27584730 Free PMC article. Review.
-
MicroRNAs derived from the insect virus HzNV-1 promote lytic infection by suppressing histone methylation.Sci Rep. 2018 Dec 13;8(1):17817. doi: 10.1038/s41598-018-35782-w. Sci Rep. 2018. PMID: 30546025 Free PMC article.
-
KSHV encoded ORF59 modulates histone arginine methylation of the viral genome to promote viral reactivation.PLoS Pathog. 2017 Jul 5;13(7):e1006482. doi: 10.1371/journal.ppat.1006482. eCollection 2017 Jul. PLoS Pathog. 2017. PMID: 28678843 Free PMC article.
References
-
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. 1995. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 86:1276–1280. - PubMed
-
- Rettig MB, Ma HJ, Vescio RA, Põld M, Schiller G, Belson D, Savage A, Nishikubo C, Wu C, Fraser J, Said JW, Berenson JR. 1997. Kaposi’s sarcoma-associated herpesvirus infection of bone marrow dendritic cells from multiple myeloma patients. Science 276:1851–1854. 10.1126/science.276.5320.1851. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA174439/CA/NCI NIH HHS/United States
- R01-CA-171979/CA/NCI NIH HHS/United States
- R01 CA171979/CA/NCI NIH HHS/United States
- R01-CA-137894/CA/NCI NIH HHS/United States
- R01-CA-1744301/CA/NCI NIH HHS/United States
- P30 DK050306/DK/NIDDK NIH HHS/United States
- R01 CA177423/CA/NCI NIH HHS/United States
- P30 CA016520/CA/NCI NIH HHS/United States
- R01-CA-177439/CA/NCI NIH HHS/United States
- P30-DK-050306/DK/NIDDK NIH HHS/United States
- R01 CA137894/CA/NCI NIH HHS/United States
- P01-CA-174439/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
