Profiling bacterial community in upper respiratory tracts
- PMID: 25391813
- PMCID: PMC4236460
- DOI: 10.1186/s12879-014-0583-3
Profiling bacterial community in upper respiratory tracts
Abstract
Background: Infection by pathogenic viruses results in rapid epithelial damage and significantly impacts on the condition of the upper respiratory tract, thus the effects of viral infection may induce changes in microbiota. Thus, we aimed to define the healthy microbiota and the viral pathogen-affected microbiota in the upper respiratory tract. In addition, any association between the type of viral agent and the resultant microbiota profile was assessed.
Methods: We analyzed the upper respiratory tract bacterial content of 57 healthy asymptomatic people (17 health-care workers and 40 community people) and 59 patients acutely infected with influenza, parainfluenza, rhino, respiratory syncytial, corona, adeno, or metapneumo viruses using culture-independent pyrosequencing.
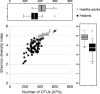
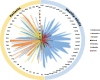
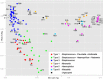
Results: The healthy subjects harbored primarily Streptococcus, whereas the patients showed an enrichment of Haemophilus or Moraxella. Quantifying the similarities between bacterial populations by using Fast UniFrac analysis indicated that bacterial profiles were apparently divisible into 6 oropharyngeal types in the tested subjects. The oropharyngeal types were not associated with the type of viruses, but were rather linked to the age of the subjects. Moraxella nonliquefaciens exhibited unprecedentedly high abundance in young subjects aged <6 years. The genome of M. nonliquefaciens was found to encode various proteins that may play roles in pathogenesis.
Conclusions: This study identified 6 oropharyngeal microbiome types. No virus-specific bacterial profile was discovered, but comparative analysis of healthy adults and patients identified a bacterium specific to young patients, M. nonliquefaciens.
Figures

Similar articles
-
The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens.Eur J Clin Microbiol Infect Dis. 2018 Sep;37(9):1725-1733. doi: 10.1007/s10096-018-3305-8. Epub 2018 Jul 22. Eur J Clin Microbiol Infect Dis. 2018. PMID: 30033505
-
Nasal microbiota dominated by Moraxella spp. is associated with respiratory health in the elderly population: a case control study.Respir Res. 2020 Jul 14;21(1):181. doi: 10.1186/s12931-020-01443-8. Respir Res. 2020. PMID: 32664929 Free PMC article.
-
Analysis of the microbiota of sputum samples from patients with lower respiratory tract infections.Acta Biochim Biophys Sin (Shanghai). 2010 Oct;42(10):754-61. doi: 10.1093/abbs/gmq081. Epub 2010 Sep 7. Acta Biochim Biophys Sin (Shanghai). 2010. PMID: 20823075
-
Viral-bacterial co-infections in the respiratory tract.Curr Opin Microbiol. 2017 Feb;35:30-35. doi: 10.1016/j.mib.2016.11.003. Epub 2016 Dec 7. Curr Opin Microbiol. 2017. PMID: 27940028 Free PMC article. Review.
-
Viral Bacterial Interactions in Children: Impact on Clinical Outcomes.Pediatr Infect Dis J. 2019 Jun;38(6S Suppl 1):S14-S19. doi: 10.1097/INF.0000000000002319. Pediatr Infect Dis J. 2019. PMID: 31205238 Free PMC article. Review.
Cited by
-
Moraxella nonliquefaciens septic arthritis in an immunocompetent child: A case report.IDCases. 2021 Apr 30;24:e01145. doi: 10.1016/j.idcr.2021.e01145. eCollection 2021. IDCases. 2021. PMID: 34026535 Free PMC article.
-
[Ocular ulcer due to Moraxella nonliquefaciens].Rev Esp Quimioter. 2019 Feb;32(1):87-88. Epub 2018 Dec 17. Rev Esp Quimioter. 2019. PMID: 30556386 Free PMC article. Spanish.
-
Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia.Front Immunol. 2018 Nov 16;9:2640. doi: 10.3389/fimmu.2018.02640. eCollection 2018. Front Immunol. 2018. PMID: 30505304 Free PMC article. Review.
-
Leveraging 3D Model Systems to Understand Viral Interactions with the Respiratory Mucosa.Viruses. 2020 Dec 11;12(12):1425. doi: 10.3390/v12121425. Viruses. 2020. PMID: 33322395 Free PMC article. Review.
-
The Effects of Smoking on Human Pharynx Microbiota Composition and Stability.Microbiol Spectr. 2023 Feb 14;11(2):e0216621. doi: 10.1128/spectrum.02166-21. Online ahead of print. Microbiol Spectr. 2023. PMID: 36786634 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

