High-throughput RNA sequencing-based virome analysis of 50 lymphoma cell lines from the Cancer Cell Line Encyclopedia project
- PMID: 25355872
- PMCID: PMC4301145
- DOI: 10.1128/JVI.02570-14
High-throughput RNA sequencing-based virome analysis of 50 lymphoma cell lines from the Cancer Cell Line Encyclopedia project
Abstract
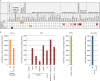
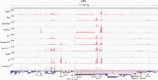
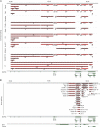
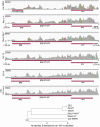
Using high-throughput RNA sequencing data from 50 common lymphoma cell culture models from the Cancer Cell Line Encyclopedia project, we performed an unbiased global interrogation for the presence of a panel of 740 viruses and strains known to infect human and other mammalian cells. This led to the findings of previously identified infections by Epstein-Barr virus (EBV), Kaposi's sarcoma herpesvirus (KSHV), and human T-lymphotropic virus type 1 (HTLV-1). In addition, we also found a previously unreported infection of one cell line (DEL) with a murine leukemia virus. High expression of murine leukemia virus (MuLV) transcripts was observed in DEL cells, and we identified four transcriptionally active integration sites, one being in the TNFRSF6B gene. We also found low levels of MuLV reads in a number of other cell lines and provided evidence suggesting cross-contamination during sequencing. Analysis of HTLV-1 integrations in two cell lines, HuT 102 and MJ, identified 14 and 66 transcriptionally active integration sites with potentially activating integrations in immune regulatory genes, including interleukin-15 (IL-15), IL-6ST, STAT5B, HIVEP1, and IL-9R. Although KSHV and EBV do not typically integrate into the genome, we investigated a previously identified integration of EBV into the BACH2 locus in Raji cells. This analysis identified a BACH2 disruption mechanism involving splice donor sequestration. Through viral gene expression analysis, we detected expression of stable intronic RNAs from the EBV BamHI W repeats that may be part of long transcripts spanning the repeat region. We also observed transcripts at the EBV vIL-10 locus exclusively in the Hodgkin's lymphoma cell line, Hs 611.T, the expression of which were uncoupled from other lytic genes. Assessment of the KSHV viral transcriptome in BCP-1 cells showed expression of the viral immune regulators, K2/vIL-6, K4/vIL-8-like vCCL1, and K5/E2-ubiquitin ligase 1 that was significantly higher than expression of the latency-associated nuclear antigen. Together, this investigation sheds light into the virus composition across these lymphoma model systems and provides insights into common viral mechanistic principles.
Importance: Viruses cause cancer in humans. In lymphomas the Epstein-Barr virus (EBV), Kaposi's sarcoma herpesvirus (KSHV) and human T-lymphotropic virus type 1 are major contributors to oncogenesis. We assessed virus-host interactions using a high throughput sequencing method that facilitates the discovery of new virus-host associations and the investigation into how the viruses alter their host environment. We found a previously unknown murine leukemia virus infection in one cell line. We identified cellular genes, including cytokine regulators, that are disrupted by virus integration, and we determined mechanisms through which virus integration causes deregulation of cellular gene expression. Investigation into the KSHV transcriptome in the BCP-1 cell line revealed high-level expression of immune signaling genes. EBV transcriptome analysis showed expression of vIL-10 transcripts in a Hodgkin's lymphoma that was uncoupled from lytic genes. These findings illustrate unique mechanisms of viral gene regulation and to the importance of virus-mediated host immune signaling in lymphomas.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
High-Throughput Sequence Analysis of Peripheral T-Cell Lymphomas Indicates Subtype-Specific Viral Gene Expression Patterns and Immune Cell Microenvironments.mSphere. 2019 Jul 10;4(4):e00248-19. doi: 10.1128/mSphere.00248-19. mSphere. 2019. PMID: 31292228 Free PMC article.
-
Distinct patterns of viral antigen expression in Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus coinfected body-cavity-based lymphoma cell lines: potential switches in latent gene expression due to coinfection.Virology. 1999 Sep 15;262(1):18-30. doi: 10.1006/viro.1999.9876. Virology. 1999. PMID: 10489337
-
Circular DNA tumor viruses make circular RNAs.Proc Natl Acad Sci U S A. 2018 Sep 11;115(37):E8737-E8745. doi: 10.1073/pnas.1811728115. Epub 2018 Aug 27. Proc Natl Acad Sci U S A. 2018. PMID: 30150410 Free PMC article.
-
[Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV, HHV-8)].Uirusu. 2010 Dec;60(2):237-45. doi: 10.2222/jsv.60.237. Uirusu. 2010. PMID: 21488336 Review. Japanese.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
Cited by
-
Primary B Lymphocytes Infected with Kaposi's Sarcoma-Associated Herpesvirus Can Be Expanded In Vitro and Are Recognized by LANA-Specific CD4+ T Cells.J Virol. 2016 Mar 28;90(8):3849-3859. doi: 10.1128/JVI.02377-15. Print 2016 Apr. J Virol. 2016. PMID: 26819313 Free PMC article.
-
Frequent infection of human cancer xenografts with murine endogenous retroviruses in vivo.Viruses. 2015 Apr 17;7(4):2014-29. doi: 10.3390/v7042014. Viruses. 2015. PMID: 25912714 Free PMC article.
-
Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines.BMC Cancer. 2018 Oct 1;18(1):940. doi: 10.1186/s12885-018-4840-5. BMC Cancer. 2018. PMID: 30285677 Free PMC article.
-
High-Throughput Sequence Analysis of Peripheral T-Cell Lymphomas Indicates Subtype-Specific Viral Gene Expression Patterns and Immune Cell Microenvironments.mSphere. 2019 Jul 10;4(4):e00248-19. doi: 10.1128/mSphere.00248-19. mSphere. 2019. PMID: 31292228 Free PMC article.
-
Expanding the conversation on high-throughput virome sequencing standards to include consideration of microbial contamination sources.mBio. 2014 Oct 28;5(6):e01989. doi: 10.1128/mBio.01989-14. mBio. 2014. PMID: 25352620 Free PMC article. No abstract available.
References
-
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. 2012. The Cancer Cell Line Encyclopedia enables predictive modeling of anticancer drug sensitivity. Nature 483:603–607. doi:10.1038/nature11003. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- P20GM103518/GM/NIGMS NIH HHS/United States
- R01CA119917/CA/NCI NIH HHS/United States
- F30CA177267/CA/NCI NIH HHS/United States
- F31 CA180449/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 CA138268/CA/NCI NIH HHS/United States
- P20 GM103518/GM/NIGMS NIH HHS/United States
- R01 AI106676/AI/NIAID NIH HHS/United States
- R01AI101046/AI/NIAID NIH HHS/United States
- F31CA180449/CA/NCI NIH HHS/United States
- F30 CA177267/CA/NCI NIH HHS/United States
- R01 AI101046/AI/NIAID NIH HHS/United States
- R01 CA119917/CA/NCI NIH HHS/United States
- R01CA138268/CA/NCI NIH HHS/United States
- R01AI106676/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

