Limited HIV infection of central memory and stem cell memory CD4+ T cells is associated with lack of progression in viremic individuals
- PMID: 25167059
- PMCID: PMC4148445
- DOI: 10.1371/journal.ppat.1004345
Limited HIV infection of central memory and stem cell memory CD4+ T cells is associated with lack of progression in viremic individuals
Abstract
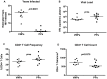
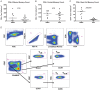
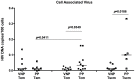
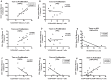
A rare subset of HIV-infected individuals, designated viremic non-progressors (VNP), remain asymptomatic and maintain normal levels of CD4+ T-cells despite persistently high viremia. To identify mechanisms potentially responsible for the VNP phenotype, we compared VNPs (average >9 years of HIV infection) to HIV-infected individuals who have similar CD4+ T-cell counts and viral load, but who are likely to progress if left untreated ("putative progressors", PP), thus avoiding the confounding effect of differences related to substantial CD4+ T cell depletion. We found that VNPs, compared to PPs, had preserved levels of CD4+ stem cell memory cells (TSCM (p<0.0001), which was associated with decreased HIV infection of these cells in VNPs (r = -0.649, p = 0.019). In addition, VNPs had decreased HIV infection in CD4+ central memory (TCM) cells (p = 0.035), and the total number of TCM cells was associated with increased proliferation of memory CD4+ T cells (r = 0.733, p = 0.01). Our results suggest that, in HIV-infected VNPs, decreased infection of CD4+ TCM and TSCM, cells are involved in preservation of CD4+ T cell homeostasis and lack of disease progression despite high viremia.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Control of HIV-1 Pathogenesis in Viremic Nonprogressors Is Independent of Gag-Specific Cytotoxic T Lymphocyte Responses.J Virol. 2018 May 29;92(12):e00346-18. doi: 10.1128/JVI.00346-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29593044 Free PMC article.
-
Delineation of Homeostatic Immune Signatures Defining Viremic Non-progression in HIV-1 Infection.Front Immunol. 2020 Mar 5;11:182. doi: 10.3389/fimmu.2020.00182. eCollection 2020. Front Immunol. 2020. PMID: 32194543 Free PMC article.
-
Higher levels of HIV DNA in memory and naive CD4(+) T cell subsets of viremic compared to non-viremic patients after 18 and 24 months of HAART.Antiviral Res. 2001 Jun;50(3):197-206. doi: 10.1016/s0166-3542(01)00142-5. Antiviral Res. 2001. PMID: 11397507
-
T memory stem cells and HIV: a long-term relationship.Curr HIV/AIDS Rep. 2015 Mar;12(1):33-40. doi: 10.1007/s11904-014-0246-4. Curr HIV/AIDS Rep. 2015. PMID: 25578055 Free PMC article. Review.
-
[Advances in the Role of Stem Memory T Cells in HIV-1 Infection].Bing Du Xue Bao. 2016 Nov;32(6):796-9. Bing Du Xue Bao. 2016. PMID: 30004654 Review. Chinese.
Cited by
-
Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection.Sci Transl Med. 2016 Sep 28;8(358):358ra125. doi: 10.1126/scitranslmed.aag1048. Sci Transl Med. 2016. PMID: 27683550 Free PMC article.
-
Slow progression of pediatric HIV associates with early CD8+ T cell PD-1 expression and a stem-like phenotype.JCI Insight. 2023 Feb 8;8(3):e156049. doi: 10.1172/jci.insight.156049. JCI Insight. 2023. PMID: 36602861 Free PMC article.
-
Cytomegalovirus and HIV Persistence: Pouring Gas on the Fire.AIDS Res Hum Retroviruses. 2017 Nov;33(S1):S23-S30. doi: 10.1089/aid.2017.0145. AIDS Res Hum Retroviruses. 2017. PMID: 29140108 Free PMC article. Review.
-
Impaired human immunodeficiency virus type 1 replicative fitness in atypical viremic non-progressor individuals.AIDS Res Ther. 2017 Mar 20;14:15. doi: 10.1186/s12981-017-0144-0. eCollection 2017. AIDS Res Ther. 2017. PMID: 28331526 Free PMC article.
-
The frequency of CD4+ and CD8+ circulating T stem cell memory in type 1 diabetes.Immun Inflamm Dis. 2022 Oct;10(10):e715. doi: 10.1002/iid3.715. Immun Inflamm Dis. 2022. PMID: 36169248 Free PMC article.
References
-
- Deeks SG, Walker BD (2007) Human Immunodeficiency Virus Controllers: Mechanisms of Durable Virus Control in the Absence of Antiretroviral Therapy. Immunity 27: 406–416. - PubMed
-
- Sodora DL, Silvestri G (2008) Immune activation and AIDS pathogenesis. AIDS 22: 439–446. - PubMed
-
- Paiardini M, Cervasi B, Dunham R, Sumpter B, Radziewicz H, et al. (2004) Cell-cycle dysregulation in the immunopathogenesis of AIDS. Immunol Res 29: 253–268. - PubMed
-
- Cohen OJ, Kinter A, Fauci AS (1997) Host factors in the pathogenesis of HIV disease. Immunol Rev 159: 31–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

