LANA binds to multiple active viral and cellular promoters and associates with the H3K4methyltransferase hSET1 complex
- PMID: 25033463
- PMCID: PMC4102568
- DOI: 10.1371/journal.ppat.1004240
LANA binds to multiple active viral and cellular promoters and associates with the H3K4methyltransferase hSET1 complex
Abstract
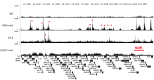
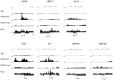
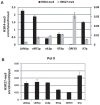
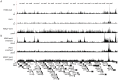
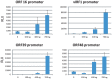
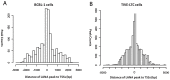
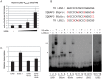
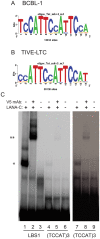
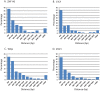
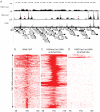
Kaposi's sarcoma-associated herpesvirus (KSHV) is a γ-herpesvirus associated with KS and two lymphoproliferative diseases. Recent studies characterized epigenetic modification of KSHV episomes during latency and determined that latency-associated genes are associated with H3K4me3 while most lytic genes are associated with the silencing mark H3K27me3. Since the latency-associated nuclear antigen (LANA) (i) is expressed very early after de novo infection, (ii) interacts with transcriptional regulators and chromatin remodelers, and (iii) regulates the LANA and RTA promoters, we hypothesized that LANA may contribute to the establishment of latency through epigenetic control. We performed a detailed ChIP-seq analysis in cells of lymphoid and endothelial origin and compared H3K4me3, H3K27me3, polII, and LANA occupancy. On viral episomes LANA binding was detected at numerous lytic and latent promoters, which were transactivated by LANA using reporter assays. LANA binding was highly enriched at H3K4me3 peaks and this co-occupancy was also detected on many host gene promoters. Bioinformatic analysis of enriched LANA binding sites in combination with biochemical binding studies revealed three distinct binding patterns. A small subset of LANA binding sites showed sequence homology to the characterized LBS1/2 sequence in the viral terminal repeat. A large number of sites contained a novel LANA binding motif (TCCAT)3 which was confirmed by gel shift analysis. Third, some viral and cellular promoters did not contain LANA binding sites and are likely enriched through protein/protein interaction. LANA was associated with H3K4me3 marks and in PEL cells 86% of all LANA bound promoters were transcriptionally active, leading to the hypothesis that LANA interacts with the machinery that methylates H3K4. Co-immunoprecipitation demonstrated LANA association with endogenous hSET1 complexes in both lymphoid and endothelial cells suggesting that LANA may contribute to the epigenetic profile of KSHV episomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Site-specific association with host and viral chromatin by Kaposi's sarcoma-associated herpesvirus LANA and its reversal during lytic reactivation.J Virol. 2014 Jun;88(12):6762-77. doi: 10.1128/JVI.00268-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696474 Free PMC article.
-
Activated Nrf2 Interacts with Kaposi's Sarcoma-Associated Herpesvirus Latency Protein LANA-1 and Host Protein KAP1 To Mediate Global Lytic Gene Repression.J Virol. 2015 Aug;89(15):7874-92. doi: 10.1128/JVI.00895-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995248 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus terminal repeat regulates inducible lytic gene promoters.J Virol. 2024 Feb 20;98(2):e0138623. doi: 10.1128/jvi.01386-23. Epub 2024 Jan 19. J Virol. 2024. PMID: 38240593 Free PMC article.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
-
The latency-associated nuclear antigen, a multifunctional protein central to Kaposi's sarcoma-associated herpesvirus latency.Future Microbiol. 2011 Dec;6(12):1399-413. doi: 10.2217/fmb.11.137. Future Microbiol. 2011. PMID: 22122438 Free PMC article. Review.
Cited by
-
Regulation of KSHV Latency and Lytic Reactivation.Viruses. 2020 Sep 17;12(9):1034. doi: 10.3390/v12091034. Viruses. 2020. PMID: 32957532 Free PMC article. Review.
-
Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication.PLoS Pathog. 2015 Jul 2;11(7):e1004993. doi: 10.1371/journal.ppat.1004993. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26133373 Free PMC article.
-
Global epigenomic analysis of KSHV-infected primary effusion lymphoma identifies functional MYC superenhancers and enhancer RNAs.Proc Natl Acad Sci U S A. 2020 Sep 1;117(35):21618-21627. doi: 10.1073/pnas.1922216117. Epub 2020 Aug 18. Proc Natl Acad Sci U S A. 2020. PMID: 32817485 Free PMC article.
-
Borna disease virus phosphoprotein modulates epigenetic signaling in neurons to control viral replication.J Virol. 2015 Jun;89(11):5996-6008. doi: 10.1128/JVI.00454-15. Epub 2015 Mar 25. J Virol. 2015. PMID: 25810554 Free PMC article.
-
KSHV Topologically Associating Domains in Latent and Reactivated Viral Chromatin.J Virol. 2022 Jul 27;96(14):e0056522. doi: 10.1128/jvi.00565-22. Epub 2022 Jul 11. J Virol. 2022. PMID: 35867573 Free PMC article.
References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. - PubMed
-
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ (1995) Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378: 505–508. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

