Influenza A and B virus intertypic reassortment through compatible viral packaging signals
- PMID: 25008914
- PMCID: PMC4178878
- DOI: 10.1128/JVI.01440-14
Influenza A and B virus intertypic reassortment through compatible viral packaging signals
Abstract
Influenza A and B viruses cocirculate in humans and together cause disease and seasonal epidemics. These two types of influenza viruses are evolutionarily divergent, and exchange of genetic segments inside coinfected cells occurs frequently within types but never between influenza A and B viruses. Possible mechanisms inhibiting the intertypic reassortment of genetic segments could be due to incompatible protein functions of segment homologs, a lack of processing of heterotypic segments by influenza virus RNA-dependent RNA polymerase, an inhibitory effect of viral proteins on heterotypic virus function, or an inability to specifically incorporate heterotypic segments into budding virions. Here, we demonstrate that the full-length hemagglutinin (HA) of prototype influenza B viruses can complement the function of multiple influenza A viruses. We show that viral noncoding regions were sufficient to drive gene expression for either type A or B influenza virus with its cognate or heterotypic polymerase. The native influenza B virus HA segment could not be incorporated into influenza A virus virions. However, by adding the influenza A virus packaging signals to full-length influenza B virus glycoproteins, we rescued influenza A viruses that possessed HA, NA, or both HA and NA of influenza B virus. Furthermore, we show that, similar to single-cycle infectious influenza A virus, influenza B virus cannot incorporate heterotypic transgenes due to packaging signal incompatibilities. Altogether, these results demonstrate that the lack of influenza A and B virus reassortants can be attributed at least in part to incompatibilities in the virus-specific packaging signals required for effective segment incorporation into nascent virions.
Importance: Reassortment of influenza A or B viruses provides an evolutionary strategy leading to unique genotypes, which can spawn influenza A viruses with pandemic potential. However, the mechanism preventing intertypic reassortment or gene exchange between influenza A and B viruses is not well understood. Nucleotides comprising the coding termini of each influenza A virus gene segment are required for specific segment incorporation during budding. Whether influenza B virus shares a similar selective packaging strategy or if packaging signals prevent intertypic reassortment remains unknown. Here, we provide evidence suggesting a similar mechanism of influenza B virus genome packaging. Furthermore, by appending influenza A virus packaging signals onto influenza B virus segments, we rescued recombinant influenza A/B viruses that could reassort in vitro with another influenza A virus. These findings suggest that the divergent evolution of packaging signals aids with the speciation of influenza A and B viruses and is in part responsible for the lack of intertypic viral reassortment.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
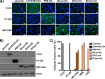
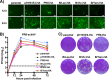

Similar articles
-
Heterologous Packaging Signals on Segment 4, but Not Segment 6 or Segment 8, Limit Influenza A Virus Reassortment.J Virol. 2017 May 12;91(11):e00195-17. doi: 10.1128/JVI.00195-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28331085 Free PMC article.
-
H5N8 and H7N9 packaging signals constrain HA reassortment with a seasonal H3N2 influenza A virus.Proc Natl Acad Sci U S A. 2019 Mar 5;116(10):4611-4618. doi: 10.1073/pnas.1818494116. Epub 2019 Feb 13. Proc Natl Acad Sci U S A. 2019. PMID: 30760600 Free PMC article.
-
Rewiring the RNAs of influenza virus to prevent reassortment.Proc Natl Acad Sci U S A. 2009 Sep 15;106(37):15891-6. doi: 10.1073/pnas.0908897106. Epub 2009 Sep 8. Proc Natl Acad Sci U S A. 2009. PMID: 19805230 Free PMC article.
-
Selective packaging of the influenza A genome and consequences for genetic reassortment.Trends Microbiol. 2014 Aug;22(8):446-55. doi: 10.1016/j.tim.2014.04.001. Epub 2014 May 2. Trends Microbiol. 2014. PMID: 24798745 Review.
-
Packaging signal of influenza A virus.Virol J. 2021 Feb 17;18(1):36. doi: 10.1186/s12985-021-01504-4. Virol J. 2021. PMID: 33596956 Free PMC article. Review.
Cited by
-
Reassortment in segmented RNA viruses: mechanisms and outcomes.Nat Rev Microbiol. 2016 Jul;14(7):448-60. doi: 10.1038/nrmicro.2016.46. Epub 2016 May 23. Nat Rev Microbiol. 2016. PMID: 27211789 Free PMC article. Review.
-
The Development and Use of Reporter Influenza B Viruses.Viruses. 2019 Aug 9;11(8):736. doi: 10.3390/v11080736. Viruses. 2019. PMID: 31404985 Free PMC article. Review.
-
Safety, Immunogenicity, and Protective Efficacy of an H5N1 Chimeric Cold-Adapted Attenuated Virus Vaccine in a Mouse Model.Viruses. 2021 Dec 3;13(12):2420. doi: 10.3390/v13122420. Viruses. 2021. PMID: 34960689 Free PMC article.
-
Implications of segment mismatch for influenza A virus evolution.J Gen Virol. 2018 Jan;99(1):3-16. doi: 10.1099/jgv.0.000989. Epub 2017 Dec 15. J Gen Virol. 2018. PMID: 29244017 Free PMC article. Review.
-
Replication-Competent ΔNS1 Influenza A Viruses Expressing Reporter Genes.Viruses. 2021 Apr 17;13(4):698. doi: 10.3390/v13040698. Viruses. 2021. PMID: 33920517 Free PMC article.
References
-
- Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

