MYC cofactors: molecular switches controlling diverse biological outcomes
- PMID: 24939054
- PMCID: PMC4143105
- DOI: 10.1101/cshperspect.a014399
MYC cofactors: molecular switches controlling diverse biological outcomes
Abstract
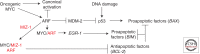
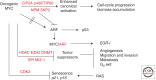
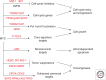
The transcription factor MYC has fundamental roles in proliferation, apoptosis, tumorigenesis, and stem cell pluripotency. Over the last 30 years extensive information has been gathered on the numerous cofactors that interact with MYC and the target genes that are regulated by MYC as a means of understanding the molecular mechanisms controlling its diverse roles. Despite significant advances and perhaps because the amount of information learned about MYC is overwhelming, there has been little consensus on the molecular functions of MYC that mediate its critical biological roles. In this perspective, the major MYC cofactors that regulate the various transcriptional activities of MYC, including canonical and noncanonical transactivation and transcriptional repression, will be reviewed and a model of how these transcriptional mechanisms control MYC-mediated proliferation, apoptosis, and tumorigenesis will be presented. The basis of the model is that a variety of cofactors form dynamic MYC transcriptional complexes that can switch the molecular and biological functions of MYC to yield a diverse range of outcomes in a cell-type- and context-dependent fashion.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

Similar articles
-
Domain-specific c-Myc ubiquitylation controls c-Myc transcriptional and apoptotic activity.Proc Natl Acad Sci U S A. 2013 Jan 15;110(3):978-83. doi: 10.1073/pnas.1208334110. Epub 2012 Dec 31. Proc Natl Acad Sci U S A. 2013. PMID: 23277542 Free PMC article.
-
TIP49, but not TRRAP, modulates c-Myc and E2F1 dependent apoptosis.Oncogene. 2002 Aug 29;21(38):5835-43. doi: 10.1038/sj.onc.1205763. Oncogene. 2002. PMID: 12185582
-
Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism.Proc Natl Acad Sci U S A. 2011 Jan 11;108(2):632-7. doi: 10.1073/pnas.1008848108. Epub 2010 Dec 27. Proc Natl Acad Sci U S A. 2011. PMID: 21187408 Free PMC article.
-
Mechanism of transcriptional activation by the Myc oncoproteins.Semin Cancer Biol. 2006 Aug;16(4):242-52. doi: 10.1016/j.semcancer.2006.08.001. Epub 2006 Aug 4. Semin Cancer Biol. 2006. PMID: 16935524 Review.
-
Myc: a weapon of mass destruction.Cell. 2004 Apr 16;117(2):153-6. doi: 10.1016/s0092-8674(04)00336-8. Cell. 2004. PMID: 15084254 Review.
Cited by
-
ChromNet: Learning the human chromatin network from all ENCODE ChIP-seq data.Genome Biol. 2016 Apr 30;17:82. doi: 10.1186/s13059-016-0925-0. Genome Biol. 2016. PMID: 27139377 Free PMC article.
-
MYC interaction with the tumor suppressive SWI/SNF complex member INI1 regulates transcription and cellular transformation.Cell Cycle. 2016 Jul 2;15(13):1693-705. doi: 10.1080/15384101.2016.1146836. Epub 2016 Jun 7. Cell Cycle. 2016. PMID: 27267444 Free PMC article.
-
Transient stabilization, rather than inhibition, of MYC amplifies extrinsic apoptosis and therapeutic responses in refractory B-cell lymphoma.Leukemia. 2019 Oct;33(10):2429-2441. doi: 10.1038/s41375-019-0454-4. Epub 2019 Mar 26. Leukemia. 2019. PMID: 30914792 Free PMC article.
-
Identification of a novel MYC target gene set signature for predicting the prognosis of osteosarcoma patients.Front Oncol. 2023 Jun 5;13:1169430. doi: 10.3389/fonc.2023.1169430. eCollection 2023. Front Oncol. 2023. PMID: 37342196 Free PMC article.
-
Biophysical and Structural Methods to Study the bHLHZip Region of Human c-MYC.Methods Mol Biol. 2021;2318:21-43. doi: 10.1007/978-1-0716-1476-1_3. Methods Mol Biol. 2021. PMID: 34019285
References
-
- Adhikary S, Eilers M 2005. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 - PubMed
-
- Alitalo K, Ramsay G, Bishop JM, Pfeifer SO, Colby WW, Levinson AD 1983. Identification of nuclear proteins encoded by viral and cellular myc oncogenes. Nature 306: 274–277 - PubMed
-
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, et al. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 7: 303–310 - PubMed
-
- Arvanitis C, Felsher DW 2006. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol 16: 313–317 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
