Lipidomics identifies a requirement for peroxisomal function during influenza virus replication
- PMID: 24868094
- PMCID: PMC4076098
- DOI: 10.1194/jlr.M049148
Lipidomics identifies a requirement for peroxisomal function during influenza virus replication
Abstract
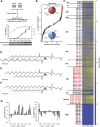
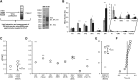
Influenza virus acquires a host-derived lipid envelope during budding, yet a convergent view on the role of host lipid metabolism during infection is lacking. Using a mass spectrometry-based lipidomics approach, we provide a systems-scale perspective on membrane lipid dynamics of infected human lung epithelial cells and purified influenza virions. We reveal enrichment of the minor peroxisome-derived ether-linked phosphatidylcholines relative to bulk ester-linked phosphatidylcholines in virions as a unique pathogenicity-dependent signature for influenza not found in other enveloped viruses. Strikingly, pharmacological and genetic interference with peroxisomal and ether lipid metabolism impaired influenza virus production. Further integration of our lipidomics results with published genomics and proteomics data corroborated altered peroxisomal lipid metabolism as a hallmark of influenza virus infection in vitro and in vivo. Influenza virus may therefore tailor peroxisomal and particularly ether lipid metabolism for efficient replication.
Keywords: biochemistry; ether lipids; glycerophospholipids; lipid metabolism; sphingolipids; systems biology.
Copyright © 2014 by the American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Oroxylin A suppresses influenza A virus replication correlating with neuraminidase inhibition and induction of IFNs.Biomed Pharmacother. 2018 Jan;97:385-394. doi: 10.1016/j.biopha.2017.10.140. Epub 2017 Nov 6. Biomed Pharmacother. 2018. PMID: 29091888
-
Ceramide Suppresses Influenza A Virus Replication In Vitro.J Virol. 2019 Mar 21;93(7):e00053-19. doi: 10.1128/JVI.00053-19. Print 2019 Apr 1. J Virol. 2019. PMID: 30700605 Free PMC article.
-
Influenza Virus with Increased pH of Hemagglutinin Activation Has Improved Replication in Cell Culture but at the Cost of Infectivity in Human Airway Epithelium.J Virol. 2019 Aug 13;93(17):e00058-19. doi: 10.1128/JVI.00058-19. Print 2019 Sep 1. J Virol. 2019. PMID: 31189708 Free PMC article.
-
Ether lipids and a peroxisomal riddle in sperm.Front Cell Dev Biol. 2023 May 15;11:1166232. doi: 10.3389/fcell.2023.1166232. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37397249 Free PMC article. Review.
-
Lipidomics Issues on Human Positive ssRNA Virus Infection: An Update.Metabolites. 2020 Aug 31;10(9):356. doi: 10.3390/metabo10090356. Metabolites. 2020. PMID: 32878290 Free PMC article. Review.
Cited by
-
Peroxisome Plasticity at the Virus-Host Interface.Trends Microbiol. 2019 Nov;27(11):906-914. doi: 10.1016/j.tim.2019.06.006. Epub 2019 Jul 19. Trends Microbiol. 2019. PMID: 31331665 Free PMC article. Review.
-
Metabolic host response and therapeutic approaches to influenza infection.Cell Mol Biol Lett. 2020 Mar 5;25:15. doi: 10.1186/s11658-020-00211-2. eCollection 2020. Cell Mol Biol Lett. 2020. PMID: 32161622 Free PMC article. Review.
-
Phospholipid analysis of two influenza A virus-infected cell lines differing in their viral replication kinetics.Arch Virol. 2023 Apr 7;168(5):132. doi: 10.1007/s00705-023-05766-x. Arch Virol. 2023. PMID: 37027089 Free PMC article.
-
Development of a High Coverage Pseudotargeted Lipidomics Method Based on Ultra-High Performance Liquid Chromatography-Mass Spectrometry.Anal Chem. 2018 Jun 19;90(12):7608-7616. doi: 10.1021/acs.analchem.8b01331. Epub 2018 Jun 8. Anal Chem. 2018. PMID: 29807422 Free PMC article.
-
Plasma lipidomics of tuberculosis patients: altered phosphatidylcholine remodeling.Future Sci OA. 2017 Oct 20;4(1):FSO255. doi: 10.4155/fsoa-2017-0011. eCollection 2018 Jan. Future Sci OA. 2017. PMID: 29255627 Free PMC article.
References
-
- Karlas A., Machuy N., Shin Y., Pleissner K. P., Artarini A., Heuer D., Becker D., Khalil H., Ogilvie L. A., Hess S., et al. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 463: 818–822 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

