Proteome analysis of subsarcolemmal cardiomyocyte mitochondria: a comparison of different analytical platforms
- PMID: 24865490
- PMCID: PMC4100094
- DOI: 10.3390/ijms15069285
Proteome analysis of subsarcolemmal cardiomyocyte mitochondria: a comparison of different analytical platforms
Abstract
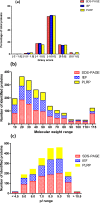
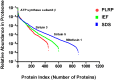
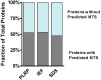
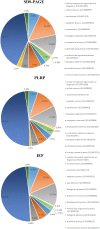
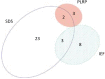
Mitochondria are complex organelles that play critical roles in diverse aspects of cellular function. Heart disease and a number of other pathologies are associated with perturbations in the molecular machinery of the mitochondria. Therefore, comprehensive, unbiased examination of the mitochondrial proteome represents a powerful approach toward system-level insights into disease mechanisms. A crucial aspect in proteomics studies is design of bioanalytical strategies that maximize coverage of the complex repertoire of mitochondrial proteins. In this study, we evaluated the performance of gel-based and gel-free multidimensional platforms for profiling of the proteome in subsarcolemmal mitochondria harvested from rat heart. We compared three different multidimensional proteome fractionation platforms: polymeric reversed-phase liquid chromatography at high pH (PLRP), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and isoelectric focusing (IEF) separations combined with liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS), and bioinformatics for protein identification. Across all three platforms, a total of 1043 proteins were identified. Among the three bioanalytical strategies, SDS-PAGE followed by LC-MS/MS provided the best coverage of the mitochondrial proteome. With this platform, 890 proteins with diverse physicochemical characteristics were identified; the mitochondrial protein panel encompassed proteins with various functional roles including bioenergetics, protein import, and mitochondrial fusion. Taken together, results of this study provide a large-scale view of the proteome in subsarcolemmal mitochondria from the rat heart, and aid in the selection of optimal bioanalytical platforms for differential protein expression profiling of mitochondria in health and disease.
Figures
Similar articles
-
Proteomics of yeast mitochondria.Methods Mol Biol. 2007;372:543-57. doi: 10.1007/978-1-59745-365-3_37. Methods Mol Biol. 2007. PMID: 18314750
-
Phosphoproteome mapping of cardiomyocyte mitochondria in a rat model of heart failure.Mol Cell Biochem. 2014 Apr;389(1-2):159-67. doi: 10.1007/s11010-013-1937-7. Epub 2014 Jan 7. Mol Cell Biochem. 2014. PMID: 24395194 Free PMC article.
-
Phosphoproteome analysis by in-gel isoelectric focusing and tandem mass spectrometry.Methods Mol Biol. 2009;519:383-96. doi: 10.1007/978-1-59745-281-6_25. Methods Mol Biol. 2009. PMID: 19381597
-
Two-dimensional electrophoresis of membrane proteins.Anal Bioanal Chem. 2007 Oct;389(4):1033-45. doi: 10.1007/s00216-007-1514-6. Epub 2007 Aug 7. Anal Bioanal Chem. 2007. PMID: 17680235 Review.
-
Recent developments and contributions from Chinese scientists in multidimensional separations for proteomics and traditional Chinese medicines.J Sep Sci. 2007 Apr;30(6):785-91. doi: 10.1002/jssc.200600372. J Sep Sci. 2007. PMID: 17536722 Free PMC article. Review.
Cited by
-
Mitochondria and ageing: role in heart, skeletal muscle and adipose tissue.J Cachexia Sarcopenia Muscle. 2017 Jun;8(3):349-369. doi: 10.1002/jcsm.12178. Epub 2017 Apr 21. J Cachexia Sarcopenia Muscle. 2017. PMID: 28432755 Free PMC article. Review.
-
Differentially expressed proteins underlying childhood cortical dysplasia with epilepsy identified by iTRAQ proteomic profiling.PLoS One. 2017 Feb 21;12(2):e0172214. doi: 10.1371/journal.pone.0172214. eCollection 2017. PLoS One. 2017. PMID: 28222113 Free PMC article.
-
Dusp26 phosphatase regulates mitochondrial respiration and oxidative stress and protects neuronal cell death.Cell Mol Life Sci. 2022 Mar 21;79(4):198. doi: 10.1007/s00018-022-04162-z. Cell Mol Life Sci. 2022. PMID: 35313355 Free PMC article.
-
Mitochondrial Kinases and the Role of Mitochondrial Protein Phosphorylation in Health and Disease.Life (Basel). 2021 Jan 23;11(2):82. doi: 10.3390/life11020082. Life (Basel). 2021. PMID: 33498615 Free PMC article. Review.
References
-
- Khan M.U., Cheema Y., Shahbaz A.U., Ahokas R.A., Sun Y., Gerling I.C., Bhattacharya S.K., Weber K.T. Mitochondria play a central role in nonischemic cardiomyocyte necrosis: Common to acute and chronic stressor states. Pflugers Arch. 2012;464:123–131. doi: 10.1007/s00424-012-1079-x. - DOI - PMC - PubMed
-
- Marin-Garcia J. Mitochondria and the Heart. Volume 256 Springer; New York, NY, USA: 2005.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

