Kaposi's sarcoma-associated herpesvirus-encoded LANA interacts with host KAP1 to facilitate establishment of viral latency
- PMID: 24741090
- PMCID: PMC4054432
- DOI: 10.1128/JVI.00596-14
Kaposi's sarcoma-associated herpesvirus-encoded LANA interacts with host KAP1 to facilitate establishment of viral latency
Abstract
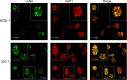
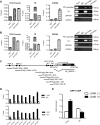
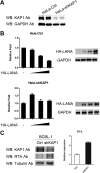
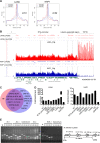
Kaposi's sarcoma-associated herpesvirus (KSHV) typically displays two different phases in its life cycle, the default latent phase and the lytic phase. There is a short period of lytic gene expression in the early stage of KSHV primary infection. The factors involved in the shutdown process of lytic gene expression are poorly identified. It has been shown that the latency-associated nuclear antigen (LANA) encoded by KSHV plays an important role in the establishment of viral latency. In screening, we identified a host protein, Krüppel-associated box domain-associated protein 1 (KAP1), that bound to LANA. We validated the interaction between LANA and KAP1 in vivo and in vitro, as well as their colocalization in the nucleus. We mapped out that LANA interacted with both the N- and C-terminal domains of KAP1. Based on the interface of LANA-KAP1 interaction determined, we proved that LANA recruited KAP1 to the RTA promoter region of the KSHV genome. We revealed that KAP1 was involved in transcriptional repression by LANA. We found multiple cooccupation sites of LANA and KAP1 on the whole KSHV genome by chromatin immunoprecipitation for sequencing (ChIP-seq) and demonstrated that LANA-recruited KAP1 played a critical role in the shutdown of lytic gene expression during the early stage of KSHV primary infection. Taken together, our data suggest that LANA interacts with KAP1 and represses lytic gene expression to facilitate the establishment of KSHV latency.
Importance: Our study revealed the mechanism of transcriptional repression by LANA during KSHV primary infection, providing new insights into the process of KSHV latency establishment.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Regulation of KSHV Latency and Lytic Reactivation.Viruses. 2020 Sep 17;12(9):1034. doi: 10.3390/v12091034. Viruses. 2020. PMID: 32957532 Free PMC article. Review.
-
Activated Nrf2 Interacts with Kaposi's Sarcoma-Associated Herpesvirus Latency Protein LANA-1 and Host Protein KAP1 To Mediate Global Lytic Gene Repression.J Virol. 2015 Aug;89(15):7874-92. doi: 10.1128/JVI.00895-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995248 Free PMC article.
-
The Latency-Associated Nuclear Antigen of Kaposi's Sarcoma-Associated Herpesvirus Inhibits Expression of SUMO/Sentrin-Specific Peptidase 6 To Facilitate Establishment of Latency.J Virol. 2017 Aug 10;91(17):e00806-17. doi: 10.1128/JVI.00806-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28615201 Free PMC article.
-
Inhibition of KAP1 enhances hypoxia-induced Kaposi's sarcoma-associated herpesvirus reactivation through RBP-Jκ.J Virol. 2014 Jun;88(12):6873-84. doi: 10.1128/JVI.00283-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696491 Free PMC article.
-
KSHV Genome Replication and Maintenance in Latency.Adv Exp Med Biol. 2018;1045:299-320. doi: 10.1007/978-981-10-7230-7_14. Adv Exp Med Biol. 2018. PMID: 29896673 Review.
Cited by
-
Epigenetic Restriction Factors (eRFs) in Virus Infection.Viruses. 2024 Jan 25;16(2):183. doi: 10.3390/v16020183. Viruses. 2024. PMID: 38399958 Free PMC article. Review.
-
Regulation of KSHV Latency and Lytic Reactivation.Viruses. 2020 Sep 17;12(9):1034. doi: 10.3390/v12091034. Viruses. 2020. PMID: 32957532 Free PMC article. Review.
-
Establishment of Tree Shrew Animal Model for Kaposi's Sarcoma-Associated Herpesvirus (HHV-8) Infection.Front Microbiol. 2021 Sep 16;12:710067. doi: 10.3389/fmicb.2021.710067. eCollection 2021. Front Microbiol. 2021. PMID: 34603235 Free PMC article.
-
Brd/BET Proteins Influence the Genome-Wide Localization of the Kaposi's Sarcoma-Associated Herpesvirus and Murine Gammaherpesvirus Major Latency Proteins.Front Microbiol. 2020 Oct 22;11:591778. doi: 10.3389/fmicb.2020.591778. eCollection 2020. Front Microbiol. 2020. PMID: 33193257 Free PMC article.
-
Methylation of KSHV vCyclin by PRMT5 contributes to cell cycle progression and cell proliferation.PLoS Pathog. 2024 Sep 10;20(9):e1012535. doi: 10.1371/journal.ppat.1012535. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39255317 Free PMC article.
References
-
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276–1280 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

