Killer-cell Immunoglobulin-like Receptor gene linkage and copy number variation analysis by droplet digital PCR
- PMID: 24597950
- PMCID: PMC4062048
- DOI: 10.1186/gm537
Killer-cell Immunoglobulin-like Receptor gene linkage and copy number variation analysis by droplet digital PCR
Abstract
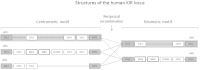
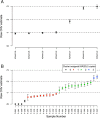
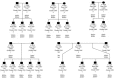
The Killer-cell Immunoglobulin-like Receptor (KIR) gene complex has considerable biomedical importance. Patterns of polymorphism in the KIR region include variability in the gene content of haplotypes and diverse structural arrangements. Droplet digital PCR (ddPCR) was used to identify different haplotype motifs and to enumerate KIR copy number variants (CNVs). ddPCR detected a variety of KIR haplotype configurations in DNA from well-characterized cell lines. Mendelian segregation of ddPCR-estimated KIR2DL5 CNVs was observed in Gambian families and CNV typing of other KIRs was shown to be accurate when compared to an established quantitative PCR method.
Figures
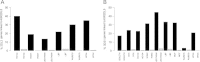

Similar articles
-
qKAT: a high-throughput qPCR method for KIR gene copy number and haplotype determination.Genome Med. 2016 Sep 29;8(1):99. doi: 10.1186/s13073-016-0358-0. Genome Med. 2016. PMID: 27686127 Free PMC article.
-
Absolute quantification reveals the stable transmission of a high copy number variant linked to autoinflammatory disease.BMC Genomics. 2016 Apr 23;17:299. doi: 10.1186/s12864-016-2619-0. BMC Genomics. 2016. PMID: 27107962 Free PMC article.
-
Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets.J Immunol. 2002 Nov 1;169(9):5118-29. doi: 10.4049/jimmunol.169.9.5118. J Immunol. 2002. PMID: 12391228
-
The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism.Immunol Rev. 2002 Dec;190:40-52. doi: 10.1034/j.1600-065x.2002.19004.x. Immunol Rev. 2002. PMID: 12493005 Review.
-
Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family.Crit Rev Immunol. 2002;22(5-6):463-82. Crit Rev Immunol. 2002. PMID: 12803322 Review.
Cited by
-
dPCR: A Technology Review.Sensors (Basel). 2018 Apr 20;18(4):1271. doi: 10.3390/s18041271. Sensors (Basel). 2018. PMID: 29677144 Free PMC article. Review.
-
Germ-line transmitted, chromosomally integrated HHV-6 and classical Hodgkin lymphoma.PLoS One. 2014 Nov 10;9(11):e112642. doi: 10.1371/journal.pone.0112642. eCollection 2014. PLoS One. 2014. PMID: 25384040 Free PMC article.
-
Identification of Novel Fusion Genes in Testicular Germ Cell Tumors.Cancer Res. 2016 Jan 1;76(1):108-16. doi: 10.1158/0008-5472.CAN-15-1790. Epub 2015 Dec 9. Cancer Res. 2016. PMID: 26659575 Free PMC article.
-
Association of Killer Cell Immunoglobulin- Like Receptor Genes in Iranian Patients with Rheumatoid Arthritis.PLoS One. 2015 Dec 11;10(12):e0143757. doi: 10.1371/journal.pone.0143757. eCollection 2015. PLoS One. 2015. PMID: 26658904 Free PMC article.
-
qKAT: a high-throughput qPCR method for KIR gene copy number and haplotype determination.Genome Med. 2016 Sep 29;8(1):99. doi: 10.1186/s13073-016-0358-0. Genome Med. 2016. PMID: 27686127 Free PMC article.
References
-
- Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little A-M, Alexander GJ, Cramp ME, O’Brien SJ, Rosenberg WMC, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. - DOI - PubMed
-
- Martin MP, Gao X, Lee J-H, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O’Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. - PubMed
-
- Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Le CT, Marsh SGE, Geraghty D, Spellman S, Haagenson MD, Ladner M, Trachtenberg E, Parham P, Miller JS. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. 2010;116:2411–2419. doi: 10.1182/blood-2010-05-283051. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

