USP11 regulates PML stability to control Notch-induced malignancy in brain tumours
- PMID: 24487962
- PMCID: PMC5645609
- DOI: 10.1038/ncomms4214
USP11 regulates PML stability to control Notch-induced malignancy in brain tumours
Erratum in
-
Erratum: USP11 regulates PML stability to control Notch-induced malignancy in brain tumours.Nat Commun. 2017 Oct 16;8:16167. doi: 10.1038/ncomms16167. Nat Commun. 2017. PMID: 29035352 Free PMC article.
Abstract
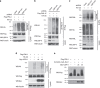
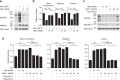
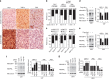
The promyelocytic leukaemia (PML) protein controls multiple tumour suppressive functions and is downregulated in diverse types of human cancers through incompletely characterized post-translational mechanisms. Here we identify USP11 as a PML regulator by RNAi screening. USP11 deubiquitinates and stabilizes PML, thereby counteracting the functions of PML ubiquitin ligases RNF4 and the KLHL20-Cul3 (Cullin 3)-Roc1 complex. We find that USP11 is transcriptionally repressed through a Notch/Hey1-dependent mechanism, leading to PML destabilization. In human glioma, Hey1 upregulation correlates with USP11 and PML downregulation and with high-grade malignancy. The Notch/Hey1-induced downregulation of USP11 and PML not only confers multiple malignant characteristics of aggressive glioma, including proliferation, invasiveness and tumour growth in an orthotopic mouse model, but also potentiates self-renewal, tumour-forming capacity and therapeutic resistance of patient-derived glioma-initiating cells. Our study uncovers a PML degradation mechanism through Notch/Hey1-induced repression of the PML deubiquitinase USP11 and suggests an important role for this pathway in brain tumour pathogenesis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures







Similar articles
-
SCP phosphatases suppress renal cell carcinoma by stabilizing PML and inhibiting mTOR/HIF signaling.Cancer Res. 2014 Dec 1;74(23):6935-46. doi: 10.1158/0008-5472.CAN-14-1330. Epub 2014 Oct 7. Cancer Res. 2014. PMID: 25293974
-
Ubiquitin-specific Protease 11 (USP11) Deubiquitinates Hybrid Small Ubiquitin-like Modifier (SUMO)-Ubiquitin Chains to Counteract RING Finger Protein 4 (RNF4).J Biol Chem. 2015 Jun 19;290(25):15526-15537. doi: 10.1074/jbc.M114.618132. Epub 2015 May 12. J Biol Chem. 2015. PMID: 25969536 Free PMC article.
-
TRIM32/USP11 Balances ARID1A Stability and the Oncogenic/Tumor-Suppressive Status of Squamous Cell Carcinoma.Cell Rep. 2020 Jan 7;30(1):98-111.e5. doi: 10.1016/j.celrep.2019.12.017. Cell Rep. 2020. PMID: 31914402
-
The role of PML ubiquitination in human malignancies.J Biomed Sci. 2012 Aug 30;19(1):81. doi: 10.1186/1423-0127-19-81. J Biomed Sci. 2012. PMID: 22935031 Free PMC article. Review.
-
PML tumour suppression and beyond: therapeutic implications.FEBS Lett. 2014 Aug 19;588(16):2653-62. doi: 10.1016/j.febslet.2014.02.007. Epub 2014 Feb 15. FEBS Lett. 2014. PMID: 24548562 Review.
Cited by
-
USP11 promotes prostate cancer progression by up-regulating AR and c-Myc activity.Proc Natl Acad Sci U S A. 2024 Jul 30;121(31):e2403331121. doi: 10.1073/pnas.2403331121. Epub 2024 Jul 25. Proc Natl Acad Sci U S A. 2024. PMID: 39052835
-
Mechanism of Notch Signaling Pathway in Malignant Progression of Glioblastoma and Targeted Therapy.Biomolecules. 2024 Apr 15;14(4):480. doi: 10.3390/biom14040480. Biomolecules. 2024. PMID: 38672496 Free PMC article. Review.
-
USP22 regulates APL differentiation via PML-RARα stabilization and IFN repression.Cell Death Discov. 2024 Mar 11;10(1):128. doi: 10.1038/s41420-024-01894-8. Cell Death Discov. 2024. PMID: 38467608 Free PMC article.
-
NORAD-Regulated Signaling Pathways in Breast Cancer Progression.Cancers (Basel). 2024 Feb 1;16(3):636. doi: 10.3390/cancers16030636. Cancers (Basel). 2024. PMID: 38339387 Free PMC article. Review.
-
USP11 promotes glycolysis by regulating HIF-1α stability in hepatocellular carcinoma.J Cell Mol Med. 2024 Jan;28(2):e18017. doi: 10.1111/jcmm.18017. Epub 2024 Jan 16. J Cell Mol Med. 2024. PMID: 38229475 Free PMC article.
References
-
- de The H., Chomienne C., Lanotte M., Degos L. & Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature 347, 558–561 (1990). - PubMed
-
- de The H. et al. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66, 675–684 (1991). - PubMed
-
- Bernardi R. & Pandolfi P. P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell. Biol. 8, 1006–1016 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

