Cell tropism predicts long-term nucleotide substitution rates of mammalian RNA viruses
- PMID: 24415935
- PMCID: PMC3887100
- DOI: 10.1371/journal.ppat.1003838
Cell tropism predicts long-term nucleotide substitution rates of mammalian RNA viruses
Abstract
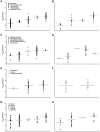
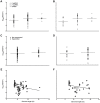
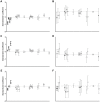
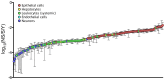
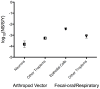
The high rates of RNA virus evolution are generally attributed to replication with error-prone RNA-dependent RNA polymerases. However, these long-term nucleotide substitution rates span three orders of magnitude and do not correlate well with mutation rates or selection pressures. This substitution rate variation may be explained by differences in virus ecology or intrinsic genomic properties. We generated nucleotide substitution rate estimates for mammalian RNA viruses and compiled comparable published rates, yielding a dataset of 118 substitution rates of structural genes from 51 different species, as well as 40 rates of non-structural genes from 28 species. Through ANCOVA analyses, we evaluated the relationships between these rates and four ecological factors: target cell, transmission route, host range, infection duration; and three genomic properties: genome length, genome sense, genome segmentation. Of these seven factors, we found target cells to be the only significant predictors of viral substitution rates, with tropisms for epithelial cells or neurons (P<0.0001) as the most significant predictors. Further, one-tailed t-tests showed that viruses primarily infecting epithelial cells evolve significantly faster than neurotropic viruses (P<0.0001 and P<0.001 for the structural genes and non-structural genes, respectively). These results provide strong evidence that the fastest evolving mammalian RNA viruses infect cells with the highest turnover rates: the highly proliferative epithelial cells. Estimated viral generation times suggest that epithelial-infecting viruses replicate more quickly than viruses with different cell tropisms. Our results indicate that cell tropism is a key factor in viral evolvability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes.Mol Biol Evol. 2004 Jun;21(6):1074-80. doi: 10.1093/molbev/msh109. Epub 2004 Mar 10. Mol Biol Evol. 2004. PMID: 15014142 Free PMC article.
-
Baculovirus Molecular Evolution via Gene Turnover and Recurrent Positive Selection of Key Genes.J Virol. 2017 Oct 27;91(22):e01319-17. doi: 10.1128/JVI.01319-17. Print 2017 Nov 15. J Virol. 2017. PMID: 28814516 Free PMC article.
-
HA-Dependent Tropism of H5N1 and H7N9 Influenza Viruses to Human Endothelial Cells Is Determined by Reduced Stability of the HA, Which Allows the Virus To Cope with Inefficient Endosomal Acidification and Constitutively Expressed IFITM3.J Virol. 2019 Dec 12;94(1):e01223-19. doi: 10.1128/JVI.01223-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597765 Free PMC article.
-
[Genome virology: the novel interaction of RNA viruses and host genomes].Uirusu. 2012 Jun;62(1):47-55. doi: 10.2222/jsv.62.47. Uirusu. 2012. PMID: 23189824 Review. Japanese.
-
RNA replication errors and the evolution of virus pathogenicity and virulence.Curr Opin Virol. 2014 Dec;9:143-7. doi: 10.1016/j.coviro.2014.09.017. Epub 2014 Oct 22. Curr Opin Virol. 2014. PMID: 25462446 Review.
Cited by
-
Animal Virus Ecology and Evolution Are Shaped by the Virus Host-Body Infiltration and Colonization Pattern.Pathogens. 2019 May 25;8(2):72. doi: 10.3390/pathogens8020072. Pathogens. 2019. PMID: 31130619 Free PMC article.
-
Zoonotic and reverse zoonotic transmission of viruses between humans and pigs.APMIS. 2021 Dec;129(12):675-693. doi: 10.1111/apm.13178. Epub 2021 Oct 18. APMIS. 2021. PMID: 34586648 Free PMC article. Review.
-
An unusually high substitution rate in transplant-associated BK polyomavirus in vivo is further concentrated in HLA-C-bound viral peptides.PLoS Pathog. 2018 Oct 18;14(10):e1007368. doi: 10.1371/journal.ppat.1007368. eCollection 2018 Oct. PLoS Pathog. 2018. PMID: 30335851 Free PMC article.
-
Why are RNA virus mutation rates so damn high?PLoS Biol. 2018 Aug 13;16(8):e3000003. doi: 10.1371/journal.pbio.3000003. eCollection 2018 Aug. PLoS Biol. 2018. PMID: 30102691 Free PMC article.
-
The Strange Lifestyle of Multipartite Viruses.PLoS Pathog. 2016 Nov 3;12(11):e1005819. doi: 10.1371/journal.ppat.1005819. eCollection 2016 Nov. PLoS Pathog. 2016. PMID: 27812219 Free PMC article. Review.
References
-
- Holmes EC (2009) The evolution and emergence of RNA viruses. Oxford: Oxford University Press. 254 p.
-
- Peters CJ (2007) Emerging viral diseases. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA et al..., editors. Fields Virology. 5th ed. Philadelphia, PA: Lippincott Williams & Wilkins. pp. 605–625.
-
- World Health Organization (2012) Vaccine-preventable diseases. Available: http://www.who.int/immunization_monitoring/diseases/en/. Accessed 20 September 2012.
-
- Rheingans RD, Antil L, Dreibelbis R, Podewils LJ, Bresee JS, et al. (2009) Economic costs of rotavirus gastroenteritis and cost-effectiveness of vaccination in developing countries. J Infect Dis 200 Suppl 1: S16–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

