Genomewide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011
- PMID: 24371052
- PMCID: PMC3958060
- DOI: 10.1128/JVI.02163-13
Genomewide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011
Abstract
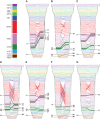
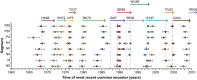
Influenza A(H3N2) viruses became widespread in humans during the 1968 H3N2 virus pandemic and have been a major cause of influenza epidemics ever since. These viruses evolve continuously by reassortment and genomic evolution. Antigenic drift is the cause for the need to update influenza vaccines frequently. Using two data sets that span the entire period of circulation of human influenza A(H3N2) viruses, it was shown that influenza A(H3N2) virus evolution can be mapped to 13 antigenic clusters. Here we analyzed the full genomes of 286 influenza A(H3N2) viruses from these two data sets to investigate the genomic evolution and reassortment patterns. Numerous reassortment events were found, scattered over the entire period of virus circulation, but most prominently in viruses circulating between 1991 and 1998. Some of these reassortment events persisted over time, and one of these coincided with an antigenic cluster transition. Furthermore, selection pressures and nucleotide and amino acid substitution rates of all proteins were studied, including those of the recently discovered PB1-N40, PA-X, PA-N155, and PA-N182 proteins. Rates of nucleotide and amino acid substitutions were most pronounced for the hemagglutinin, neuraminidase, and PB1-F2 proteins. Selection pressures were highest in hemagglutinin, neuraminidase, matrix 1, and nonstructural protein 1. This study of genotype in relation to antigenic phenotype throughout the period of circulation of human influenza A(H3N2) viruses leads to a better understanding of the evolution of these viruses.
Importance: Each winter, influenza virus infects approximately 5 to 15% of the world's population, resulting in significant morbidity and mortality. Influenza A(H3N2) viruses evolve continuously by reassortment and genomic evolution. This leads to changes in antigenic recognition (antigenic drift) which make it necessary to update vaccines against influenza A(H3N2) viruses frequently. In this study, the relationship of genetic evolution to antigenic change spanning the entire period of A(H3N2) virus circulation was studied for the first time. The results presented in this study contribute to a better understanding of genetic evolution in correlation with antigenic evolution of influenza A(H3N2) viruses.
Figures
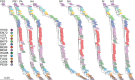

Similar articles
-
Antigenic drift and subtype interference shape A(H3N2) epidemic dynamics in the United States.Elife. 2024 Sep 25;13:RP91849. doi: 10.7554/eLife.91849. Elife. 2024. PMID: 39319780 Free PMC article.
-
Predominance of influenza A(H3N2) viruses during the 2016/2017 season in Bulgaria.J Med Microbiol. 2018 Feb;67(2):228-239. doi: 10.1099/jmm.0.000668. Epub 2018 Jan 3. J Med Microbiol. 2018. PMID: 29297852
-
Genetic analysis of influenza A/H3N2 and A/H1N1 viruses circulating in Vietnam from 2001 to 2006.J Clin Microbiol. 2008 Feb;46(2):399-405. doi: 10.1128/JCM.01549-07. Epub 2007 Oct 17. J Clin Microbiol. 2008. PMID: 17942644 Free PMC article.
-
[Molecular characterization of human influenza viruses--a look back on the last 10 years].Berl Munch Tierarztl Wochenschr. 2006 Mar-Apr;119(3-4):167-78. Berl Munch Tierarztl Wochenschr. 2006. PMID: 16573207 Review. German.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
Cited by
-
A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies.Nat Commun. 2021 Mar 19;12(1):1722. doi: 10.1038/s41467-021-21954-2. Nat Commun. 2021. PMID: 33741916 Free PMC article.
-
Genetic Lineage and Reassortment of Influenza C Viruses Circulating between 1947 and 2014.J Virol. 2016 Aug 26;90(18):8251-65. doi: 10.1128/JVI.00969-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27384661 Free PMC article.
-
RNA Sequence Features Are at the Core of Influenza A Virus Genome Packaging.J Mol Biol. 2019 Oct 4;431(21):4217-4228. doi: 10.1016/j.jmb.2019.03.018. Epub 2019 Mar 23. J Mol Biol. 2019. PMID: 30914291 Free PMC article. Review.
-
Implications of segment mismatch for influenza A virus evolution.J Gen Virol. 2018 Jan;99(1):3-16. doi: 10.1099/jgv.0.000989. Epub 2017 Dec 15. J Gen Virol. 2018. PMID: 29244017 Free PMC article. Review.
-
The evolution of seasonal influenza viruses.Nat Rev Microbiol. 2018 Jan;16(1):47-60. doi: 10.1038/nrmicro.2017.118. Epub 2017 Oct 30. Nat Rev Microbiol. 2018. PMID: 29081496 Review.
References
-
- Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814–2822. 10.1128/JVI.79.5.2814-2822.2005 - DOI - PMC - PubMed
-
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274. 10.1073/pnas.1116200109 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

