Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus
- PMID: 24336184
- PMCID: PMC5708296
- DOI: 10.1038/nrmicro3161
Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus
Abstract
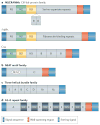
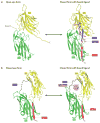
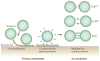
Staphylococcus aureus is an important opportunistic pathogen and persistently colonizes about 20% of the human population. Its surface is 'decorated' with proteins that are covalently anchored to the cell wall peptidoglycan. Structural and functional analysis has identified four distinct classes of surface proteins, of which microbial surface component recognizing adhesive matrix molecules (MSCRAMMs) are the largest class. These surface proteins have numerous functions, including adhesion to and invasion of host cells and tissues, evasion of immune responses and biofilm formation. Thus, cell wall-anchored proteins are essential virulence factors for the survival of S. aureus in the commensal state and during invasive infections, and targeting them with vaccines could combat S. aureus infections.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Cell Wall-Anchored Surface Proteins of Staphylococcus aureus: Many Proteins, Multiple Functions.Curr Top Microbiol Immunol. 2017;409:95-120. doi: 10.1007/82_2015_5002. Curr Top Microbiol Immunol. 2017. PMID: 26667044
-
The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation.Microbiology (Reading). 2007 Aug;153(Pt 8):2435-2446. doi: 10.1099/mic.0.2007/006676-0. Microbiology (Reading). 2007. PMID: 17660408
-
Cell-wall-anchored proteins affect invasive host colonization and biofilm formation in Staphylococcus aureus.Microbiol Res. 2024 Aug;285:127782. doi: 10.1016/j.micres.2024.127782. Epub 2024 May 27. Microbiol Res. 2024. PMID: 38833832 Review.
-
Surface proteins and the formation of biofilms by Staphylococcus aureus.Biochim Biophys Acta Biomembr. 2018 Mar;1860(3):749-756. doi: 10.1016/j.bbamem.2017.12.003. Epub 2017 Dec 9. Biochim Biophys Acta Biomembr. 2018. PMID: 29229527 Free PMC article.
-
The MSCRAMM Family of Cell-Wall-Anchored Surface Proteins of Gram-Positive Cocci.Trends Microbiol. 2019 Nov;27(11):927-941. doi: 10.1016/j.tim.2019.06.007. Epub 2019 Jul 30. Trends Microbiol. 2019. PMID: 31375310 Review.
Cited by
-
Toolbox of Characterized Genetic Parts for Staphylococcus aureus.ACS Synth Biol. 2024 Jan 19;13(1):103-118. doi: 10.1021/acssynbio.3c00325. Epub 2023 Dec 8. ACS Synth Biol. 2024. PMID: 38064657 Free PMC article.
-
Phenol-soluble modulin α induces G2/M phase transition delay in eukaryotic HeLa cells.FASEB J. 2015 May;29(5):1950-9. doi: 10.1096/fj.14-260513. Epub 2015 Feb 3. FASEB J. 2015. PMID: 25648996 Free PMC article.
-
The biophysics of bacterial infections: Adhesion events in the light of force spectroscopy.Cell Surf. 2021 Jan 15;7:100048. doi: 10.1016/j.tcsw.2021.100048. eCollection 2021 Dec. Cell Surf. 2021. PMID: 33665520 Free PMC article.
-
Antibiotic Resistance Profiles and MLST Typing of Staphylococcus Aureus Clone Associated with Skin and Soft Tissue Infections in a Hospital of China.Infect Drug Resist. 2024 Jun 21;17:2555-2566. doi: 10.2147/IDR.S465951. eCollection 2024. Infect Drug Resist. 2024. PMID: 38933775 Free PMC article.
-
Non-antibiotic strategies for prevention and treatment of internalized Staphylococcus aureus.Front Microbiol. 2022 Aug 31;13:974984. doi: 10.3389/fmicb.2022.974984. eCollection 2022. Front Microbiol. 2022. PMID: 36118198 Free PMC article. Review.
References
-
- Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu Rev Microbiol. 2010;64:143–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

